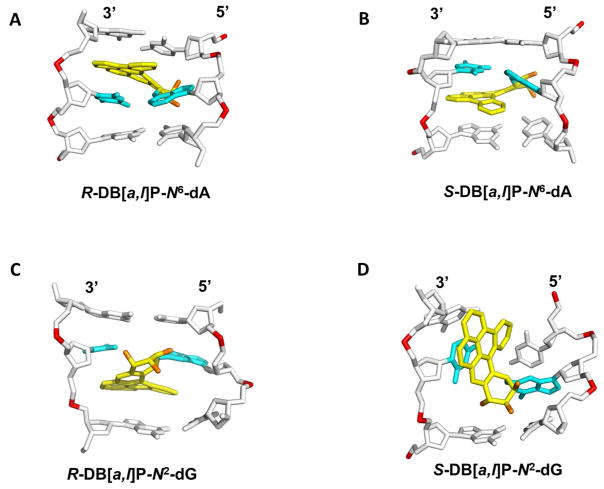
Figure 4.
Best representative structures from MD simulations for the (A) R and (B) S-DB[a,l]P-N6-dA 58 adducts and NMR solution structures for the (C) R-DB[a,l]P-N2-dG 59 and (D) S-DB[a,l]P-N2-dG 60 adducts. The central trimers of the duplex 11-mers are shown. The full duplex 11-mers are shown in Supporting Information Figure S6. The view is looking into the minor groove. For the DB[a,l]P moiety, the carbon atoms are colored yellow and the oxygen atoms orange. The damaged base pairs are colored cyan and the DNA duplexes are colored white, except for the phosphorus atoms, which are colored red. Hydrogen atoms are not displayed for clarity. Coordinates for (D) are provided in Supporting Information Table S1.