Figure 1.
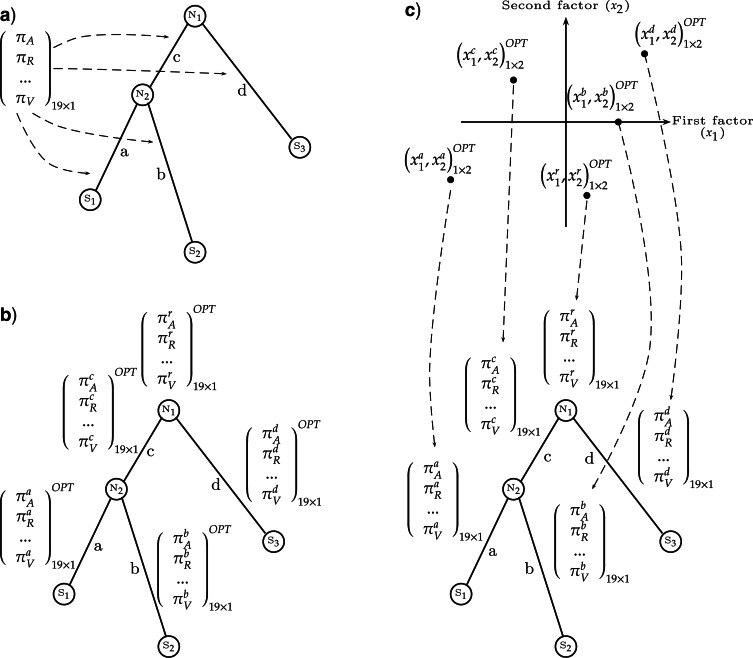
The COaLA model substantially decreases the dimension of the space of equilibrium frequency parameters. a) In homogeneous and stationary models, only one vector of amino acid frequencies represents the equilibrium state of sequences and is used for likelihood computation. This vector may be optimized by ML (LG+Fopt model) or not (LG or LG + Fobs models). b) With a standard nonhomogeneous approach, the homogeneity and stationarity hypotheses are relaxed by assigning independent vectors of 19 equilibrium frequencies per branch to model the variations of overall composition through time. c) With the COaLA model, small dimension vectors of coordinates along the first axes of the COA are optimized per branch. In this example, a two-dimension vector corresponding to the first two axes is associated to each branch and is optimized by ML (OPT). Reversing the COA (dashed arrows), from a vector of coordinates in the low-dimension space, one can compute the corresponding vector of 20 frequencies that is used to compute transition probabilities along the branch.