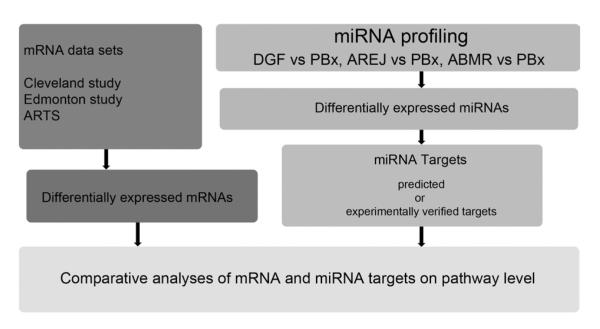
FIGURE 2.
Schematic representation of our combinatorial approach for identifying miRNAs, miRNA targets, genes, and molecular pathways in DGF and acute rejection. Differentially expressed miRNAs were obtained by comparing DGF, AREJ, and ABMR compared with allografts with normal function (PBx). miRNA target genes were predicted by DIANAmT, miRanda, and Targetscan (42–44), and experimentally validated targets were identified by using miRTarBase (45). Posttransplantation gene expression profiles were derived from the Cleveland study (2) and from the Edmonton study (6). ARTS was obtained from analysis of an independent gene expression data set of renal allograft biopsies with acute rejection (8). Comparison of miRNA target lists and gene lists were performed to derive set of pathways core to acute rejection and DGF. ABMR, antibody-mediated rejection; AREJ, acute cellular rejection; ARTS, acute rejection transcript set; DGF, delayed graft function; miRNA, microRNA; PBx, protocol biopsy.