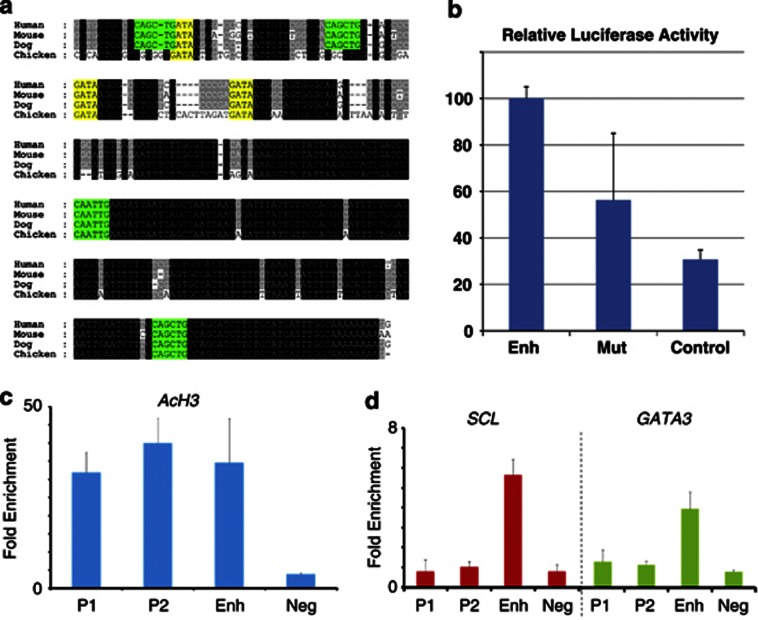
Figure 6.
SCL/TAL1 and GATA3 bind to the LMO1 +57 enhancer in T-ALL patient cells. (a) Nucleotide sequence alignment of the LMO1 gene (+57 kb) enhancer region. Alignment of human, mouse, dog and chicken sequences extracted from the UCSC genome browser with conserved GATA (yellow) and E-Box (green) motifs coloured for clarity. Black boxes indicate 100% cross-species sequence conservation, grey boxes show less conserved sequences. (b) Stable transfection assays in Jurkat cells show that deletion of the region containing conserved E-Box and GATA motifs causes a significant reduction in activity of the +57 enhancer. Data from four technical replicates each of three biological replicates were normalised against the wild-type enhancer. The results show the mean of these data points with error bars representing s.d. Paired t-test analysis using all 12 raw data-points for each construct confirmed that the reduction in enhancer activity seen with the mutant enhancer was highly significant (P<0.01). (c) ChIP assays using an antibody against acetylated histone H3 (H3K9/K14) reveal elevated levels of histone acetylation at both LMO1 promoters as well as the +57 enhancer region in an LMO1-expressing primagraft primary patient sample. (d) ChIP assays on T-ALL cells from the same primagraft (X31) using antibodies against SCL/TAL1and GATA3. Analysis by quantitative PCR demonstrates significant binding of both SCL/TAL1 and GATA3 to the LMO1 +57 enhancer, with no binding to either of the two LMO1 promoters.