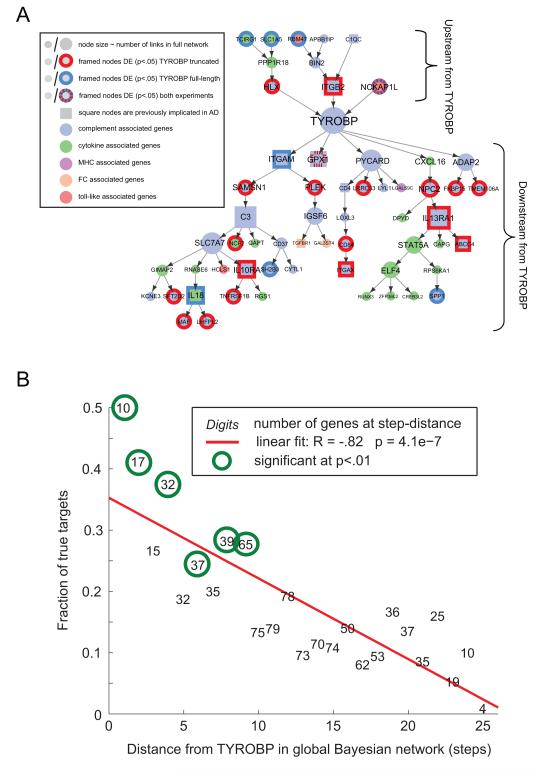
Figure 6. Structure of Causal Networks Guides Differential Expression in a Distance-dependent Manner.
A) Within the microglia module, we show all genes which receive direct or indirect causal inputs to/from TYROBP. Genes which were differentially expressed in either full-length or truncated Tyrobp experiments are circled (P-value<0.05, n=4/4/4 for control/truncated/full-length RNA sequenced samples). Possible reasons for differentially expressed (DE) of predicted upstream genes are mouse-human network differences, network inaccuracy, or presence of feedback loops, which are not represented in a Bayesian framework. (B) We mapped results of RNA sequencing experiments onto a large Bayesian network of ~8000 nodes that contains the microglia module as well as many other modules. In this large network, we could track differential expression of genes which are predicted to be downstream of TYROBP at various network distances (link distances). There was a strong negative correlation (r= −0.82, P=4.e-07) between the differentially expressed genes in the microglia and the path distance from TYROBP in the brain immune network. See also Figure S6 and Table S1.