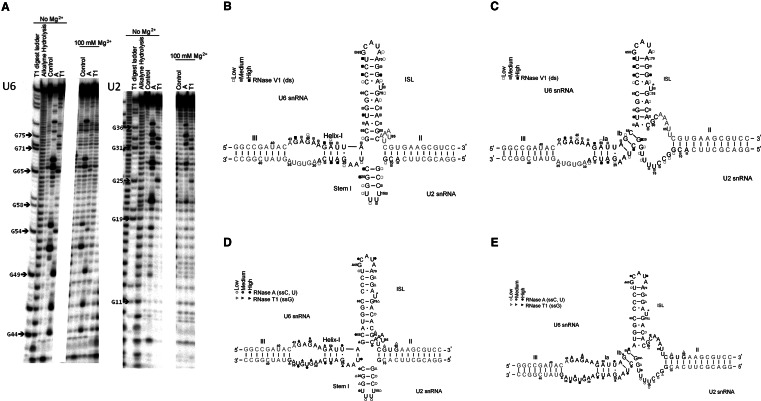
FIGURE 2.
(A) Representative sequencing gel exhibiting cleavage patterns of 32P-labeled (*)hU2 and *hU6 snRNA paired with respective counter-strands subjected to enzymatic probing by ribonucleases RNase A and T1 (see Materials and Methods for experimental details). Lanes are labeled on top with the ribonuclease used and the concentration of Mg2+. (B,C) Normalized cleavage intensities of complex between *hU6 and hU2 snRNAs and the complex between *hU2 and hU6 snRNAs following reaction with RNase V1 were mapped onto the two possible folds (B: four-helix model; C: three-helix model) of the U2-U6 snRNA complex. Cleavage by RNase V1 at the corresponding nucleotides is represented by squares, with open, gray, and black squares corresponding to low, medium, and high intensity, respectively. (D,E) Normalized cleavage intensities of complex between *hU6 and hU2 snRNAs and the complex between *hU2 and hU6 snRNAs following reaction with RNases A and T1 were mapped onto the two folds as in Figure 2, B and C. Cleavage by RNase A and RNase T1 at corresponding nucleotides is represented by open, gray, and black circles or triangles, respectively, corresponding to low, medium, and high cleavage by the ribonuclease, respectively. Nucleotides in gray represent those for which information was not collected because of gel artifacts.