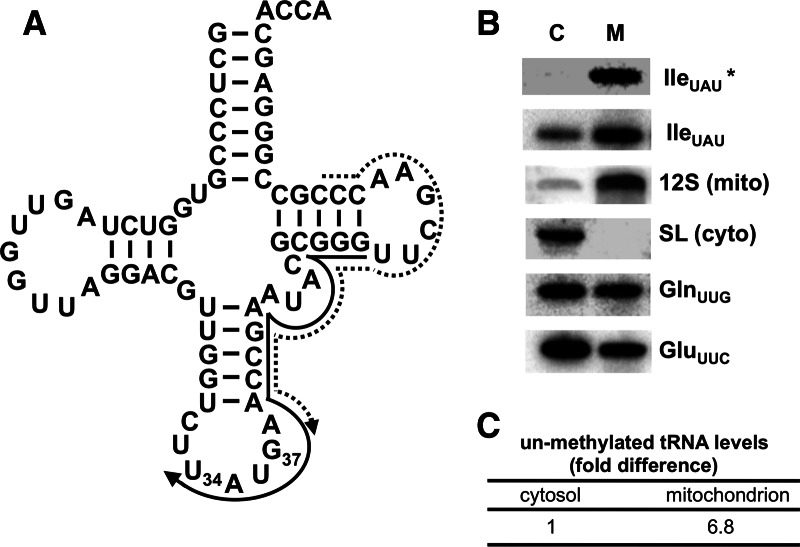
FIGURE 1.
Presence of m1G37-lacking tRNAIle UAU in mitochondria. (A) Cloverleaf structure of tRNAIle UAU. U34 corresponds to the first position of the anticodon, and G37 is the site of m1G formation. The solid arrow represents the G37-overlapping oligonucleotide primer specific for tRNAIle UAU used as a probe in Northern hybridizations. The presence of m1G prevents hybridization of this probe. The dashed arrow represents a m1G37-nonoverlapping probe, which should not discriminate between m1G37-containing and unmethylated tRNAs. (B) Northern blots of cytosolic (C, extra-mitochondrial) and mitochondrial (M) RNA fractions. The panel shows a significant signal with the m1G37-overlapping probe, which detects nonmethylated tRNAIle UAU (IleUAU*). The probe that does not cover the methylated position shows a signal for cytoplasmic and mitochondrial fractions (IleUAU). Probes for 12S rRNA (12S, a mitochondrial marker) and spliced leader RNA (SL, cytoplasmic marker) were used as controls for fraction purity. Probes specific for tRNAGln UUG (GlnUUG) and tRNAGlu UUC (GluUUC) were used as hybridization controls for tRNAs that do not contain m1G. These probes have similar melting temperatures for hybridization and hybridize to a similar region of these tRNAs as the tRNAIle probe. (C) The relative levels of unmethylated tRNA, in either cellular fraction, were calculated by normalizing the signals for the unmethylated (IleUAU*) and the nondiscriminating probe (IleUAU) from each compartment to the tRNAGln UUG and tRNAGlu UUC signals. The normalized unmethylated value for each compartment was then divided by the total signal (from IleUAU) and the cytoplasmic level set at one. The normalized unmethylated signal for the mitochondrial fraction was divided by the normalized unmethylated cytoplasmic signal.