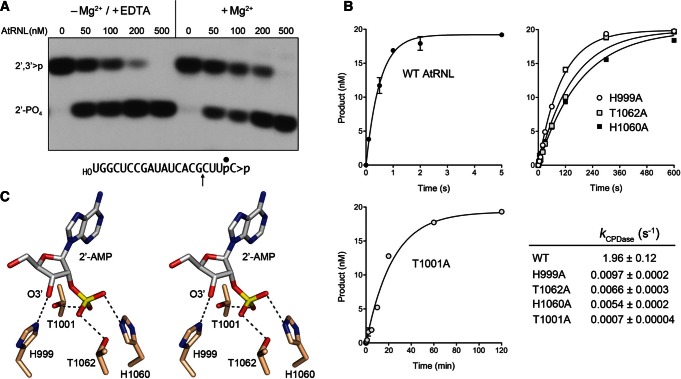
FIGURE 2.
Kinetics and mechanism of the CPD reaction. (A) Metal independence. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 2 mM DTT, 20 nM HORNA>p substrate (shown at bottom with the position of the 32P-label denoted by • and the site of RNase T1 scission indicated by ↑), increasing concentrations of AtRNL as specified, and either 10 mM EDTA or 10 mM MgCl2 were incubated for 1 min at 22°C. The products were digested with RNase T1 and then analyzed by urea-PAGE. An autoradiogram of the gel is shown. The phosphorylation states of the terminal RNase T1 fragments HOCUUpC>p (2′,3′>p) or HOCUUpC2′p (2′-PO4) are indicated at left. (B) Single-turnover kinetics. Reaction mixtures contained 20 nM HORNA>p substrate and 1 µM wild-type or mutant AtRNL as specified. The extents of cyclic phosphate hydrolysis (HOCUUpC2′p/[HOCUUpC>p + HOCUUpC2′p]) normalized to input RNA concentration are plotted as a function of reaction time. Each datum in the graphs is the average of three separate experiments ±SEM. Nonlinear regression curve fits of the data to a single exponential are shown. The CPD rate constants for the wild-type and mutant enzymes are tabulated. (C) Stereo view of the active site of mammalian 2′,3′-cyclic phosphodiesterase in complex with the reaction product 2′-AMP (from pdb 2YDD). The histidine and threonine residues are numbered according to their counterparts in AtRNL. Hydrogen bonds to the 2′-PO4 and 3′-OH are indicated by dashed lines.