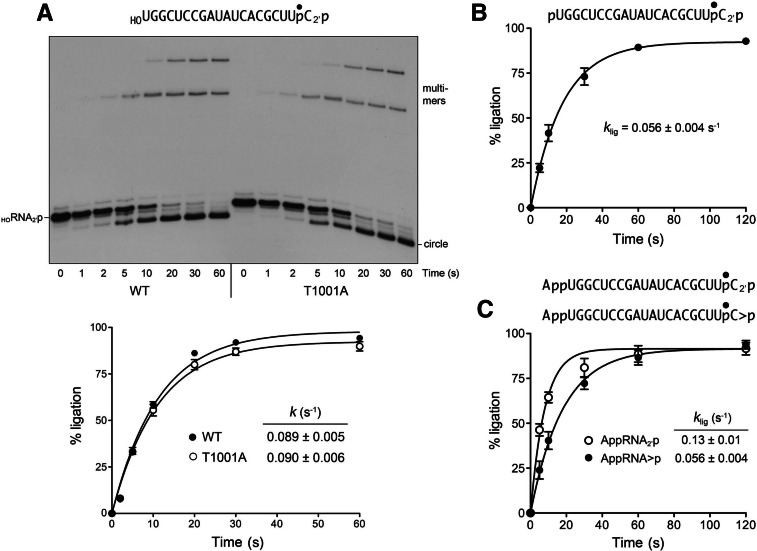
FIGURE 4.
Ligation of RNAs with pre-healed 2′-PO4 ends. (A) Kinetics of HORNA2′p ligation and bypass of the T1001A CPD mutation. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 0.1 mM ATP, 2 mM DTT, 20 nM HORNA2′p substrate (as shown), and 1 µM wild-type AtRNL or T1001A mutant as specified were incubated at 22°C. The reactions were quenched at the times indicated and the products analyzed by urea-PAGE (top panel). The extents of ligation by wild-type (WT) and T1001A AtRNL are plotted as a function of reaction time (bottom panel). Each datum is the average of four separate experiments ±SEM. Nonlinear regression curve fits of the data to a single exponential are shown. (B) Kinetics of pRNA2′p ligation. Reaction mixtures contained 20 nM pRNA2′p substrate (as shown) and 1 µM wild-type AtRNL. Each datum in the graph is the average of four separate experiments ±SEM. (C) Kinetics of AppRNA2′p ligation. Reaction mixtures contained 20 nM AppRNA2′p or AppRNA>p substrates (as shown) and 1 µM wild-type AtRNL. Each datum in the graph is the average of either four (AppRNA2′p) or five (AppRNA>p) separate time-course experiments ±SEM.