FIGURE 1.
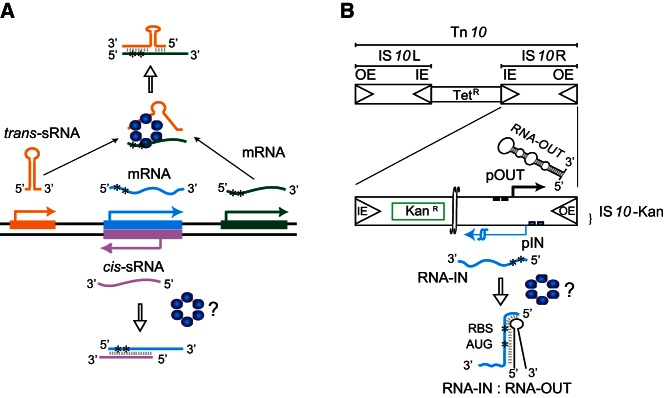
Small regulatory RNAs (sRNAs) and the Tn10/IS10 antisense system. (A) Cis- vs. trans-encoded sRNAs. Transcribed strands of three different genes and their corresponding RNAs (color coded) are shown. Pairing of a trans-sRNA (gold) and an mRNA (green) and of a cis-sRNA (pink) and an mRNA (cyan) is shown. Hfq (blue hexamer) catalyzes pairing in the former case where there is partial sequence complementarity between partners, but it is unclear if it also catalyzes pairing in the latter case where there is perfect sequence complementarity between partners. Asterisks (*) define the translation initiation region (TIR) of the mRNAs. (B) Structure of Tn10 and IS10-Kan. Tn10 is a 9147-bp composite transposon that confers tetracycline resistance (TetR). Tn10 is comprised of IS10-Left and IS10-Right, the latter of which encodes a functional transposase protein that catalyzes DNA cleavage and joining events involving the “outside” (OE) and “inside” (IE) ends. The transposase mRNA (RNA-IN) is encoded from the promoter pIN (blue squares). A second promoter (pOUT–black squares) within IS10-Right encodes a cis-sRNA (also referred to as an antisense RNA), RNA-OUT. To follow transposition of IS10-Right in E. coli, a KanR gene cassette was cloned into IS10-Right, creating IS10-Kan. RNA-OUT is depicted as a stable stem–loop structure (black) and RNA-IN is depicted as a blue line with asterisks defining the TIR. RNA-OUT is known to pair with RNA-IN, and this inhibits translation of RNA-IN, thereby down-regulating transposition. Hfq can enhance the rate of RNA-IN:OUT pairing in vitro, but it is not known if Hfq plays a role in this antisense system in vivo.
