Abstract
Peroxisomes (microbodies) are ubiquitous single-membrane–bounded organelles and fulfill essential roles in the cellular metabolism. They are found in virtually all eukaryotic cells and basically multiply by division. However, the mechanochemical machinery involved in peroxisome division remains elusive. Here, we first identified the peroxisome-dividing (POD) machinery. We isolated the POD machinery from Cyanidioschyzon merolae, a unicellular red alga containing a single peroxisome. Peroxisomal division in C. merolae can be highly synchronized by light/dark cycles and the microtubule-disrupting agent oryzalin. By proteomic analysis based on the complete genome sequence of C. merolae, we identified a dynamin-related protein 3 (DRP3) ortholog, CmDnm1 (Dnm1), that predominantly accumulated with catalase in the dividing-peroxisome fraction. Immunofluorescence microscopy demonstrated that Dnm1 formed a ring at the division site of the peroxisome. The outlines of the isolated dynamin rings were dimly observed by phase-contrast microscopy and clearly stained for Dnm1. Electron microscopy revealed that the POD machinery was formed at the cytoplasmic side of the equator. Immunoelectron microscopy showed that the POD machinery consisted of an outer dynamin-based ring and an inner filamentous ring. Down-regulation of Dnm1 impaired peroxisomal division. Surprisingly, the same Dnm1 serially controlled peroxisomal division after mitochondrial division. Because genetic deficiencies of Dnm1 orthologs in multiperoxisomal organisms inhibited both mitochondrial and peroxisomal proliferation, it is thought that peroxisomal division by contraction of a dynamin-based machinery is universal among eukaryotes. These findings are useful for understanding the fundamental systems in eukaryotic cells.
Peroxisomes are single-membrane–bounded organelles found in nearly all eukaryotic cells. In plant cells, peroxisomes are involved in a variety of metabolic pathways essential for development associated with photorespiration, lipid mobilization, and hormone biosynthesis (1, 2). In animals, abnormalities in peroxisome proliferation are associated with carcinogenesis, neurodegeneration, and cerebrohepatorenal syndrome (1, 3). Peroxisomes are thought to basically proliferate by division, although they do not contain DNA (1). Because the cells of multiperoxisomal organisms, such as yeasts, plants, and animals, contain irregularly shaped peroxisomes that divide randomly, their proliferation has been examined by analyzing peroxisome abundance and distribution (4, 5). Therefore, the division machinery (ring) that is essential for proliferation and plays a central role is unclear. Cyanidioschyzon merolae offers unique advantages for studying peroxisomal division, because each cell contains a minimal set of basic eukaryotic organelles, comprising one chloroplast, one mitochondrion, one cell nucleus, and one peroxisome, the divisions of which occur in that order and can be synchronized by light/dark cycles (6–9) (Fig. 1 A and B and Fig. S1). In C. merolae, peroxisomes do not form de novo from the endoplasmic reticulum in the peroxisomal division cycle but divide by binary fission (6, 7, 10). In addition, the complete sequence of the genome has enabled proteomic analyses (7, 11).
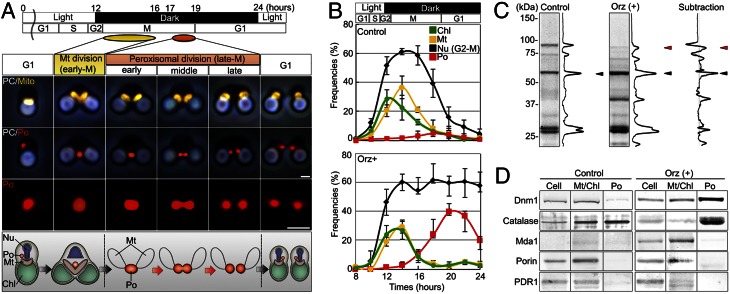
Fig. 1.
Identification of Dnm1 from the dividing-peroxisome fraction. (A) Immunofluorescence and schematic images of mitochondrial and peroxisomal divisions of C. merolae. Peroxisomal (red) division occurred after mitochondrial (yellow) division. Chl, chloroplast; Mt, mitochondrion; Nu, nucleus; PC, phase-contrast image; Po, peroxisome. (B) Frequencies of dividing cell nuclei (Nu), dividing chloroplasts (Chl), dividing mitochondria (Mt), and dividing peroxisomes (Po) in non–oryzalin-treated cells (control) and oryzalin-treated cells (Orz+) at the indicated times after synchronization (n > 100). (C) Proteomic analysis of peroxisomal fractions in control and oryzalin-treated cells at 20 h after synchronization. The major bands specific to oryzalin-treated cells were identified as catalase (black arrowhead), Dnm1 (red arrowhead), and others. (D) Immunoblot analyses of Dnm1, catalase, mitochondria division protein (Mda1), porin, and chloroplast division protein (PDR1). Cell, whole cell; Mt/Chl, isolated mitochondria and chloroplast; Po, isolated peroxisomes. (Scale bars: 1 μm.)
Results and Discussion
Proteomic Analysis of Isolated Dividing Peroxisomes.
To search for proteins essential for peroxisomal division, we developed a protocol to isolate dividing peroxisomes and analyzed them by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). Peroxisomal division occurs during late mitotic (M) phase (6, 12) (Fig. 1A and Fig. S1). After mitochondrial division during early M phase, oval-shaped peroxisomes become dumbbell-shaped and finally divide into two daughter peroxisomes by binary fission (6, 12) (Fig. 1A and Fig. S1). Because the peroxisomes divide within a very short time (12), few dividing peroxisomes were detected, even during late M phase (Fig. 1B, Upper). We previously developed a method to collect cells containing dividing peroxisomes by oryzalin treatment (12, 13). This resulted in the temporary accumulation of cells containing dividing peroxisomes (Fig. 1B and Fig. S1 A and B). Isolated peroxisomal fractions were obtained from non–oryzalin-treated (control) and oryzalin-treated cells (Fig. S2 C and D) and examined by MALDI-TOF-MS. As a result, proteins in 10 (control cells) and 13 (oryzalin-treated cells) bands were identified (Fig. 1C, Fig. S2E, and Table S1). The major subtracting bands included catalase and CmDnm1/DRP3 (Dnm1) (Fig. 1C and Fig. S2F). Dnm1 is one of only two dynamins (Dnm1 and Dnm2) encoded in the C. merolae genome (7, 11). Dnm1 and Dnm2 are important components of the mitochondrion-dividing (MD) machinery (9, 14, 15) and plastid-dividing (PD) machinery (16, 17), respectively. Immunoblot analysis indicated that Dnm1 was concentrated in the dividing-peroxisome fraction with catalase (Fig. 1D), suggesting the interesting possibility that the same Dnm1 serially controls peroxisomal division after mitochondrial division.
Subcellular Localization of Dnm1 and Isolation of Dynamin Rings.
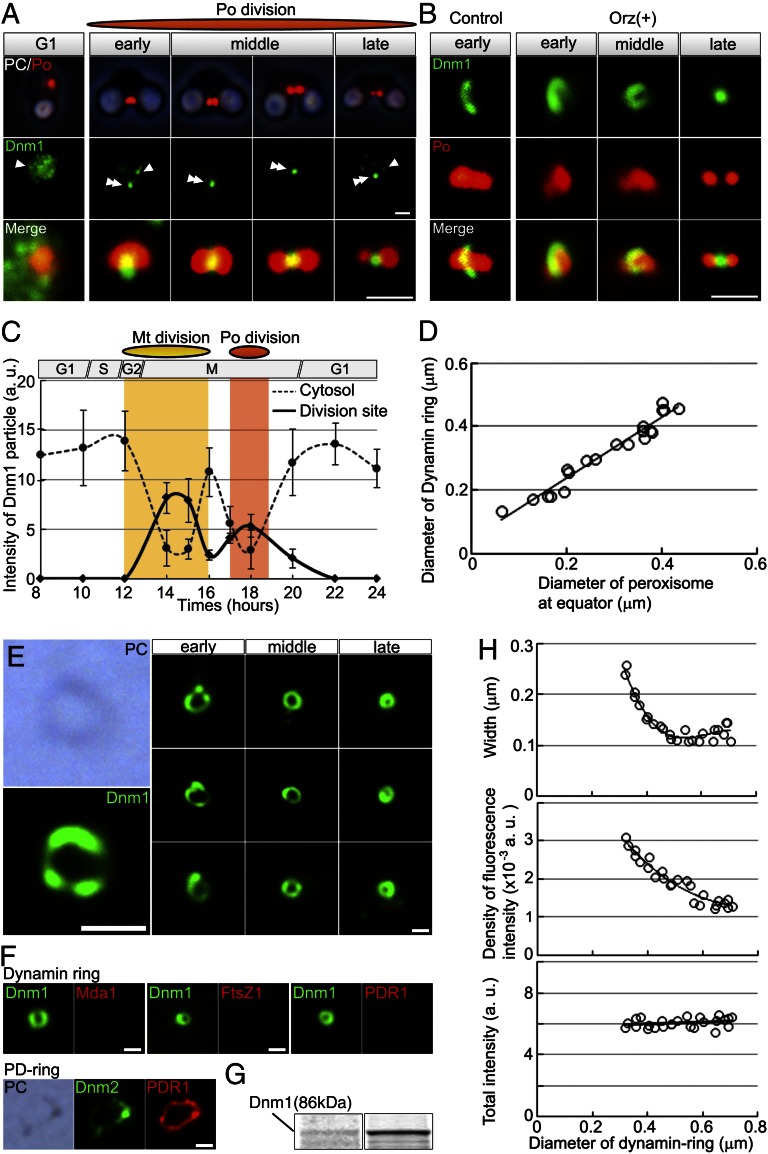
Consistent with the results of the proteomic analysis, immunofluorescence microscopy with antibodies against Dnm1 and catalase revealed that Dnm1 began to move from the cytosol to the equator of the oval-shaped peroxisome at 1.5 h after mitochondrial division, and then most Dnm1 became localized at the division site (Fig. 2 A and C). In slightly squashed oryzalin-treated and control cells, we observed dynamin ring-like structures encircling the equator of dividing peroxisomes (Fig. 2B). After separation of the daughter peroxisomes, the Dnm1 signals moved to the cytosol (Fig. 2 A and C). Similar intracellular migration of Dnm1 signals between the cytosol and organelles was observed during mitochondrial division (9) (Fig. 2C). The diameter of the dynamin ring decreased as the diameter of the division site decreased during peroxisomal division (Fig. 2D), suggesting that the dynamin ring has a role in contraction at the division site. When the bulk peroxisomal contents were dissolved using detergents (SI Text), isolated dynamin rings were clearly observed by immunostaining for Dnm1. The outlines of the dynamin rings were dimly visible by phase-contrast microscopy (Fig. 2E). The dynamin rings were not stained for the antibodies against mitochondrial-division apparatus 1 (Mda1), filamentous-tempreture sensitive Z1 (FtsZ1) or plastid-dividing ring 1 (PDR1) (Fig. 2F). SDS/PAGE and MALDI-TOF-MS analyses showed that the Dnm1 band was concentrated in the insoluble fraction (Fig. 2G). The width and density of the fluorescence intensity of the dynamin ring increased as peroxisomal division progressed, but the total fluorescence intensity of the ring did not change (Fig. 2H). These findings suggested that the peroxisome divides by contraction of the dynamin ring formed at an early period.
Fig. 2.
Dynamin localization at the peroxisome division site and isolation of dynamin rings. (A) Phase-contrast and immunofluorescence images of the peroxisome (Po) (red) and Dnm1 (green) and enlarged merged images. (B) Immunofluorescence images of the dynamin ring-like structures (green) encircling dividing peroxisomes (red) in slightly squashed control and oryzalin-treated cells. (C) Dynamic change of Dnm1 signals between the cytosol (dotted line) and division site of mitochondrion or peroxisome (solid line) during mitochondrial and peroxisomal divisions (n > 50). (D) The diameter of the dynamin ring decreases as the diameter of the division site decreases during the peroxisome division process. (E) Phase-contrast and immunofluorescence images of closed dynamin rings (green) isolated from peroxisomes at various stages of division. (F) Immunofluorescence images of isolated dynamin rings from dividing peroxisomal fraction showing Dnm1 (green) and Mda1, FtsZ1, or PDR1 (red). Images of isolated PD ring showing Dnm2 (green) and PDR1 (red) as a control. (G) SDS/PAGE of the dividing-peroxisome fraction (Left) and isolated dynamin ring fraction (Right). (H) Width, density of intensity, and total fluorescence intensity of the Dnm1 signals relative to the diameter of the dynamin ring. White arrowheads, cytosolic dynamin; double arrowhead, division site. [Scale bars: 1 μm (A and B); 500 nm (E and F).]
Composition of the POD Machinery.
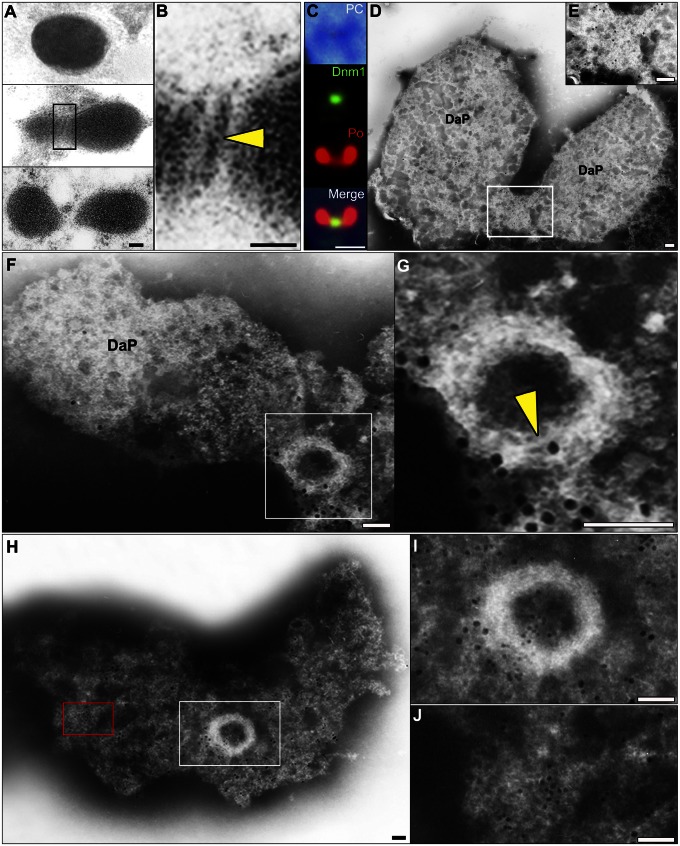
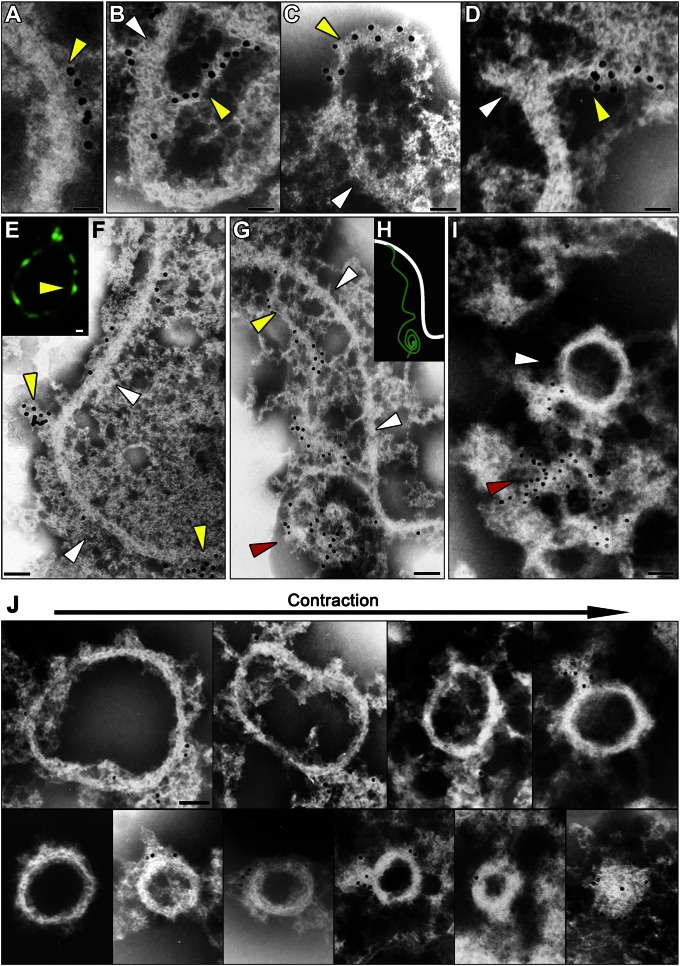
An in vitro guanosine triphosphatase (GTPase) assay did not induce a conformational change in the dynamin rings isolated from dividing peroxisomes (Fig. S3). This finding can be explained by considering the structural analogy to the PD machinery, in which motive force for the contraction of the putative ring working together in addition to the GTPase activity of dynamin in vivo is required for contraction of the dynamin ring (18). Usually, the putative ring cannot be visualized by thin-section electron microscopy (EM) after conventional fixation (6, 10). Rarely, a narrow edge of the ring was observed at the cytoplasmic side after high-pressure freeze-fixation (Fig. 3 A and B). Immunofluorescence microscopy and whole-mount EM revealed that the isolated dividing peroxisomes visualized were swollen compared with the thin-section image, and that Dnm1 was localized on the bridge between the daughter peroxisomes (Fig. 3 C–E and Fig. S4 A and B). When the membrane was partially dissolved by detergent, skeletal filamentous rings appeared (Fig. 3 H and I) with peroxisome membrane, which was labeled with catalase (Fig. 3J and Table 1). These consisted of a bundle of fine filaments of 4–5 nm in diameter (Fig. 3 F–I and Fig. S4 C–G). Dynamin-bound immunogold particles (yellow arrowheads) were arranged at distances of about 10–20 nm along the skeletal filamentous rings (Fig. 4A and Fig. S5). Dynamin localized on strings peeled away from the skeletal filamentous ring, suggesting that the POD machinery consisted of the dynamin-based ring (a complex of dynamin and amorphous string) and the filamentous ring (Fig. 4 B–I). The separated dynamin rings formed clumps (yellow arrowheads) on the filamentous rings (Fig. 4 E and F). The dynamin-based ring separated from the filamentous ring showed a string and part of the ring formed a clump, suggesting that the dynamin-based ring encircled the filamentous ring (Fig. 4 G–I). The POD machinery was more similar to the PD machinery than to the MD machinery, which formed an irregularly shaped, clumped structure (Figs. S5 C–E and S6 B and C). However, the diameter of the POD machinery was about half that of the PD machinery, and it did not form a supertwisted structure like the PD machinery (18) (Fig. S6C and Table S2). During contraction, the dynamin-based ring encircled the filamentous ring and the POD machinery decreased in diameter but its width was unchanged (Fig. 4J and Fig. S6D). Finally, the filamentous ring disappeared, leaving only the dynamin-based ring (Fig. 4J). These observations suggested that the contraction of the POD machinery was caused by sliding of the dynamin-based ring and the filamentous ring accompanied by partial disassembly.
Fig. 3.
Identification of the POD machinery in C. merolae cells by thin-section EM and immuno-EM. (A and B) Thin-section EM images showing the progress of peroxisomal division. The edge of the POD machinery was visualized as a fine bar on the cytoplasmic side of the single membrane (yellow arrowhead) at the peroxisome division site. (C) Immunofluorescence images showing Dnm1 (green) localized at the division plane of isolated peroxisomes (red). (D and E) Immuno-EM images showing a parallel image to the division plane. POD machinery containing Dnm1 was visible on bridges between the daughter peroxisomes. (F and G) Perpendicular image to the division plane. A filamentous ring (yellow arrowhead) appears after dissociation of the membrane. (H and I) Dnm1 is localized on the periphery of the POD machinery, and catalase is localized on the membrane. (J) Enlarged micrograph of the red boxed area in H showing partially dissolved peroxisome membrane labeled with catalase. Detergent treatment for 30 s (C–I); DaP, daughter peroxisome; large immunogold particles (15 nm), Dnm1 (C–J); small immunogold particles (10 nm), catalase (H–J). B, E, G, and I show enlarged micrographs of the boxed areas in A, D, F, and H, respectively. [Scale bars: 50 nm (A and B, F–H); 1μm (C); 10nm (D and E).]
Table 1.
Quantitative calculation of immunolabeling density at each site against catalase and Dnm1
| Site* | Catalase (10 nm) | Dnm1 (15 nm) |
| Membrane | 433.9 ± 62.1 | 15.0 ± 14.1 |
| POD-ring | 44.2 ± 17.3 | 304.0 ± 45.9 |
Labeling density was measured per square micron. Values are indicated as means ± SD of the number of gold particles.
Examined sites (n = 10) are listed as membrane (partially dissolved peroxisome membrane, excluding POD-ring) and POD-ring.
Fig. 4.
Immuno-EM and immunofluorescence microscopic images and schematic representation showing the separation of the filamentous ring and the dynamin-based ring. A–D, F, G, I, and J and E and H show immuno-EM and fluorescence microscopic images, respectively. (A) Part of the dynamin-based ring (yellow arrowheads) is arranged at distances of about 10 nm along the filamentous ring (white arrowheads) in the POD machinery. (B–D) The dynamin-based rings (yellow arrowheads) are peeled off from the filamentous ring (white arrowheads) in parts of the POD machinery. (E and F) The separated dynamin rings form clumps (yellow arrowheads) on the filamentous ring (white arrowheads). (G) The dynamin-based ring separated from the filamentous ring (white arrowheads) shows a string (yellow arrowheads) and a part of the ring forms a clump (red arrowhead), suggesting that the dynamin-based ring encircles the filamentous ring. (H) The schematic representation shows the dynamin-based ring (green line) and the filamentous ring (white line). (I) The dynamin-based ring separates from the filamentous ring (white arrowhead). (J) Isolated POD machinery arranged in order of division. Immunogold particles (15 nm), Dnm1 (A–D, F, and G); green, anti-Dnm1 antibody (E). [Scale bars: 50 nm (A–I); 100 nm (J).]
Down-Regulation of Dnm1 During Peroxisomal and Mitochondrial Divisions.
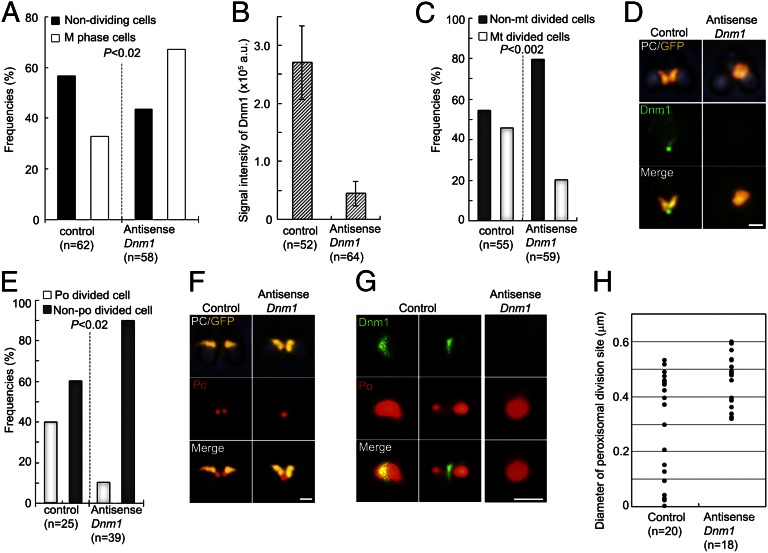
To investigate the role of dynamin in the POD machinery during peroxisomal division, we down-regulated Dnm1 using an antisense technique. The antisense-Dnm1 DNA was introduced into cells in nonsynchronized culture using a polyethylene glycol method (14). The frequency of M phase cells increased after the introduction of the antisense-Dnm1 DNA (Fig. 5A). The transformed M phase cells with down-regulated Dnm1 showed decreased signal intensity for Dnm1 (Fig. 5B) and inhibition of mitochondrial division during early M phase (Fig. 5 C and D). In addition, peroxisomal division, which occurred during late M phase, was inhibited and nondividing peroxisomes were accumulated (Fig. 5 E and F). In these cells, the dynamin ring did not appear and the peroxisomes became oval-shaped (Fig. 5 G and H). These findings supported the notion that Dnm1 plays an important role in the contraction of the POD machinery, in addition to its role in mitochondrial division.
Fig. 5.
Down-regulation of Dnm1. (A) Frequencies of nondividing cells (interphase) and M phase cells. Nondividing cells are decreased, whereas M phase cells are increased, at 48 h after introduction of the antisense-Dnm1 DNA (P < 0.02 by Fisher’s exact test). (B) The immunofluorescence intensity of the dynamin ring is decreased by the down-regulation. Data are means and SD (n > 50). (C) Down-regulation of Dnm1 inhibits mitochondrial division (n > 50; P < 0.002 by Fisher’s exact test). (D) Phase-contrast and immunofluorescence images showing a dividing mitochondrion (GFP, yellow, anti-GFP) and Dnm1 (green) in control cells and a nondivided mitochondrion without dynamin in antisense-Dnm1 DNA-treated cells. (E) Cells with down-regulated Dnm1 show inhibited peroxisomal division, and the number of non–peroxisome-dividing cells is increased during late M phase (P < 0.02 by Fisher’s exact test). Data are the frequencies of control and antisense-Dnm1 DNA-treated cells with dividing peroxisomes (po) and nondividing peroxisomes (n > 25). (F) Phase-contrast and immunofluorescence images showing a dividing mitochondrion with a dividing peroxisome in control cells and a dividing mitochondrion with a nondividing peroxisome in antisense-Dnm1 DNA-treated cells. (G) Immunofluorescence images show a dividing peroxisome (red) with a dynamin ring (green) in control cells and a nondividing oval-shaped peroxisome without a dynamin ring in Dnm1-down-regulated cells. (H) Peroxisomal division in Dnm1–down-regulated cells stops at the early stage of contraction of the POD machinery. The data represent the diameters of the peroxisome division sites in control cells and antisense-Dnm1 DNA-treated cells. (Scale bars: 1 μm.)
Function of the POD Machinery and Comparisons Between the POD Machinery and the MD/PD Machineries.
To discuss the mechanisms of peroxisomal division, the POD machinery was compared with the MD (9, 14, 15) and PD (16–18) machineries (Table S2). There were at least five differences between single- and double-membrane–bounded organelle divisions. First, the width and diameter of the POD machinery were smaller than those of the MD and PD machineries (Fig. S6). Second, the POD machinery lacked bacterium-derived dividing proteins such as FtsZ (Fig. 2F) but contained dynamin derived from host eukaryotes. Therefore, division of the single-membrane–bounded peroxisome did not require the formation of an inner ring on the matrix side. This finding may give insights into peroxisome origins. Third, although the MD and POD machineries both contain Dnm1, the isolated POD machinery contains a rigid filamentous ring like the isolated PD machinery but unlike the isolated MD machinery, which forms an irregularly shaped, clumped ring. Whereas the filamentous ring in the PD machinery consists of a bundle of PDR1-mediated polyglucans (17), the POD machinery did not contain PDR1 (Fig. 2F). The components of the filamentous ring remain to be identified. Fourth, the isolated POD machinery did not form a supertwisted ring (Figs. 3 F–I and Figs. 4 J and Figs. S5 and S6), whereas the isolated PD machinery did (Fig. S6C). Fifth, the width of the filamentous ring in the POD machinery remained constant during peroxisome contraction, whereas those of the MD and PD machineries increased during contraction (8, 15) (Figs. S6). We suggest that dynamin functions in three steps in the operation of the POD machinery (Fig. S7). First, Dnm1 molecules form a ring on the cytosolic side of the outer filamentous ring from cytoplasmic patches during the early period of peroxisomal division. Second, the dynamin-based ring becomes wider, and the filamentous ring partially disassembles during the contraction of the POD machinery. In the PD machinery, the dynamin molecules generate a contraction force by sliding the filamentous ring (18). In the POD machinery, down-regulation of Dnm1 inhibited the peroxisome contraction, suggesting that the dynamin-based ring generates the motive force for contraction of the PD machinery, as well as the POD machinery. Finally, the dynamin-based ring directly pinches off the single membrane of the bridge between the daughter peroxisomes after disassembly of the filamentous ring. The divided peroxisomes are separated into daughter cells by electron-dense connectors linking the peroxisomes and mitochondria (6, 10).
Molecular genetic studies of multiperoxisomal organisms have suggested that dynamin-like protein DRP3 orthologs are involved in both mitochondrial division (19) and peroxisomal proliferation (1, 5, 20–24). In these organisms, dynamin molecules are recruited to the both peroxisomal and mitochondrial membrane by the interactions of mitochondrial fission 1 (Fis1) (25, 26). In C. merolae, Fis1-like proteins have not been identified. Because the Dnm1 is continuously expressed throughout the cell cycle, including organelle division cycles in C. merolae (27, 28), an unknown factor that can recruit Dnm1 molecules to the division sites may play a role for the key regulator in the divisions of peroxisome and mitochondrion. Although the dynamin family members are phylogenetically grouped according to their functions, Dnm1 is grouped in the same subclade of DRP3s, which have dual functions (29, 30). Thus, serial control of peroxisomal division and mitochondrial division by the same dynamin might be conserved among virtually all eukaryotes.
Materials and Methods
Algal Materials.
We used C. merolae 10D (11).
Synchronization and Isolation of Dividing Peroxisomes.
For synchronization, cells were subjected to light/dark cycles at 42 °C (7, 18) and treated with 40 mM oryzalin for 12 h as described previously (12, 13). The method for isolation of dividing peroxisomes (SI Text) was developed from a previously described method (18).
Proteomic Analysis of the Dividing-Peroxisome Fraction.
Samples were analyzed by a peptide mass fingerprinting search using a MALDI-TOF-MS (AXIMA-TOF2; Shimadzu), based on the FASTA file distributed by the C. merolae Genome Project (7, 11).
Immunoblot Analysis and Immunofluorescence Microscopy.
For immunoblot analyses, we used an anti-Dnm1 rabbit antibody (1:1,000 dilution), anti-catalase antibody (1:2,000 dilution), anti-porin antibody (1:1,000 dilution), anti-Mda1 antibody (1:1,000 dilution), and anti-PDR1 antibody (1:500 dilution). For immunofluorescence microscopy, the secondary antibodies were Alexa Fluor 488- or Alexa Fluor 555-conjugated goat anti-rabbit, anti-mouse, or anti-rat IgG (1:1,000 dilution). Images were captured using a fluorescence microscope (BX51; Olympus).
Isolation of the POD Machinery.
The isolated dividing peroxisomes were dissolved in membrane dissolution buffer and incubated at 4 °C for 10 min. The fraction was then dissolved in purification buffer and incubated at 4 °C for 30 s. Isolation of the PD machinery rings was performed as described previously (18).
EM and Immuno-EM.
Thin-section EM was performed as described previously (17) after high-pressure freeze fixation. For immuno-EM, we used anti-Dnm1 (1:10 dilution) and anti-catalase (1:200 dilution) primary antibodies. As secondary antibodies, we used goat anti-rabbit IgG conjugated with 15-nm colloidal gold (1:10 dilution) and goat anti-rat IgG conjugated with 10-nm colloidal gold (1:10 dilution). The samples were dissolved in purification buffer, negatively stained with phosphotungstic acid, and examined using an EM (JEOL 1200).
Plasmid Construction and Transformation.
The antisense Dnm1 suppression plasmid was constructed as described in SI Text and introduced into C. merolae cells using a polyethylene glycol method (17).
Supplementary Material
Acknowledgments
This work was supported, in part, by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 22247007 and 22657061 (to T.K.); Japan Society for the Promotion of Science Fellowship 6759 (to Y.I.); and the Human Frontier Science Program Long Term Fellowship (to Y.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303483110/-/DCSupplemental.
References
- 1.Schrader M, Yoon Y. Mitochondria and peroxisomes: Are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays. 2007;29(11):1105–1114. doi: 10.1002/bies.20659. [DOI] [PubMed] [Google Scholar]
- 2.Pan R, Hu J. The conserved fission complex on peroxisomes and mitochondria. Plant Signal Behav. 2011;6(6):870–872. doi: 10.4161/psb.6.6.15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gould SJ, Valle D. Peroxisome biogenesis disorders: Genetics and cell biology. Trends Genet. 2000;16(8):340–345. doi: 10.1016/s0168-9525(00)02056-4. [DOI] [PubMed] [Google Scholar]
- 4.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J Cell Biol. 2007;178(3):399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koch A, Schneider G, Lüers GH, Schrader M. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci. 2004;117(Pt 17):3995–4006. doi: 10.1242/jcs.01268. [DOI] [PubMed] [Google Scholar]
- 6.Miyagishima S, et al. Microbody proliferation and segregation cycle in the single microbody-alga Cyanidioschyzon merolae. Planta. 1999;208(3):326–336. [Google Scholar]
- 7.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428(6983):653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 8.Miyagishima S, Itoh R, Toda K, Kuroiwa H, Kuroiwa T. Real-time analyses of chloroplast and mitochondrial division and differences in the behavior of their dividing rings during contraction. Planta. 1999;207(3):343–353. [Google Scholar]
- 9.Nishida K, et al. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci USA. 2003;100(4):2146–2151. doi: 10.1073/pnas.0436886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroiwa T, et al. Vesicle, mitochondrial, and plastid division machineries with emphasis on dynamin and electron-dense rings. Int Rev Cell Mol Biol. 2008;271:97–152. doi: 10.1016/S1937-6448(08)01203-3. [DOI] [PubMed] [Google Scholar]
- 11.Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imoto Y, et al. Division of cell nuclei, mitochondria, plastids, and microbodies mediated by mitotic spindle poles in the primitive red alga Cyanidioschyzon merolae. Protoplasma. 2010;241(1-4):63–74. doi: 10.1007/s00709-010-0107-y. [DOI] [PubMed] [Google Scholar]
- 13.Nishida K, Yagisawa F, Kuroiwa H, Nagata T, Kuroiwa T. Cell cycle-regulated, microtubule-independent organelle division in Cyanidioschyzon merolae. Mol Biol Cell. 2005;16(5):2493–2502. doi: 10.1091/mbc.E05-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida Y, et al. The bacterial ZapA-like protein ZED is required for mitochondrial division. Curr Biol. 2009;19(17):1491–1497. doi: 10.1016/j.cub.2009.07.035. [DOI] [PubMed] [Google Scholar]
- 15.Nishida K, Yagisawa F, Kuroiwa H, Yoshida Y, Kuroiwa T. WD40 protein Mda1 is purified with Dnm1 and forms a dividing ring for mitochondria before Dnm1 in Cyanidioschyzon merolae. Proc Natl Acad Sci USA. 2007;104(11):4736–4741. doi: 10.1073/pnas.0609364104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyagishima SY, et al. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell. 2003;15(3):655–665. doi: 10.1105/tpc.009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida Y, et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science. 2010;329(5994):949–953. doi: 10.1126/science.1190791. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida Y, et al. Isolated chloroplast division machinery can actively constrict after stretching. Science. 2006;313(5792):1435–1438. doi: 10.1126/science.1129689. [DOI] [PubMed] [Google Scholar]
- 19.Bleazard W, et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1(5):298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuravi K, et al. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J Cell Sci. 2006;119(Pt 19):3994–4001. doi: 10.1242/jcs.03166. [DOI] [PubMed] [Google Scholar]
- 21.Fujimoto M, et al. Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 2009;58(3):388–400. doi: 10.1111/j.1365-313X.2009.03786.x. [DOI] [PubMed] [Google Scholar]
- 22.Koch A, et al. Dynamin-like protein 1 is involved in peroxisomal fission. J Biol Chem. 2003;278(10):8597–8605. doi: 10.1074/jbc.M211761200. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Gould SJ. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J Biol Chem. 2003;278(19):17012–17020. doi: 10.1074/jbc.M212031200. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka A, Kobayashi S, Fujiki Y. Peroxisome division is impaired in a CHO cell mutant with an inactivating point-mutation in dynamin-like protein 1 gene. Exp Cell Res. 2006;312(9):1671–1684. doi: 10.1016/j.yexcr.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 25.Zhang XC, Hu JP. FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol Plant. 2008;1(6):1036–1047. doi: 10.1093/mp/ssn056. [DOI] [PubMed] [Google Scholar]
- 26.Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16(11):5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara T, et al. Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2009;16(1):59–72. doi: 10.1093/dnares/dsn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imoto Y, Yoshida Y, Yagisawa F, Kuroiwa H, Kuroiwa T. The cell cycle, including the mitotic cycle and organelle division cycles, as revealed by cytological observations. J Electron Microsc (Tokyo) 2011;60(Suppl 1):S117–S136. doi: 10.1093/jmicro/dfr034. [DOI] [PubMed] [Google Scholar]
- 29.Praefcke GJK, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5(2):133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 30.Miyagishima SY, Kuwayama H, Urushihara H, Nakanishi H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc Natl Acad Sci USA. 2008;105(39):15202–15207. doi: 10.1073/pnas.0802412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.