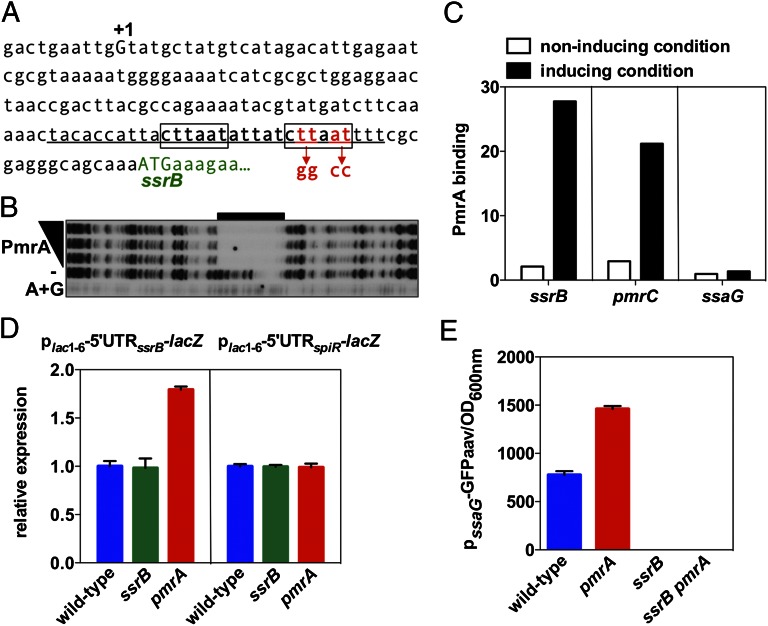
Fig. 4.
PmrA binds to the promoter of the ssrB gene. (A) DNA sequence of the promoter region of the ssrB gene. A +1 corresponds to the reported transcriptional start site, the boxed sequences indicate those matching the PmrA-binding consensus sequence, and the underlined sequence indicates the region protected by the PmrA protein in the ssrB promoter shown in B. Mutated nucleotides at the PmrA binding site in the ssrB promoter mutant strain are indicated beneath the sequence. (B) DNase I footprinting analysis of the ssrB promoter using purified PmrA-His6 protein (0, 100, 200, and 300 pmoles) was performed as described in Materials and Methods. Lane A+G corresponds to dideoxy chain-termination sequences for the ssrB promoter DNA. The black bar indicates the region protected by the PmrA-His6 protein. (C) In vivo occupancy of the ssrB, pmrC, and ssaG promoter regions by the PmrA-HA protein when Salmonella strains expressing PmrA-HA from its original chromosomal location and promoter were switched from noninducing conditions (N-minimal medium at pH 7.7 with 1 mM Mg2+) into noninducing or inducing conditions (N-minimal medium at pH 7.7 with 10 μM Mg2+ and 100 μM Fe3+). (D) β-galactosidase activity expressed by wild-type, ssrB, and pmrA Salmonella harboring a plasmid with a plac1–6-driven transcriptional fusion of the 5′ untranslated region (5′UTR) of ssrB or spiR to a promoterless lacZ gene. Bacteria were grown in N-minimal medium at pH 7.7 with 10 μM Mg2+. Values are normalized by that of the wild-type strain. (E) Fluorescence originating from a plasmid-linked ssaG-gfp transcriptional fusion was determined in wild-type, pmrA, ssrB, and ssrB pmrA Salmonella grown in N-minimal medium at pH 4.6 with 1 mM Mg2+ (LH) for 8 h.