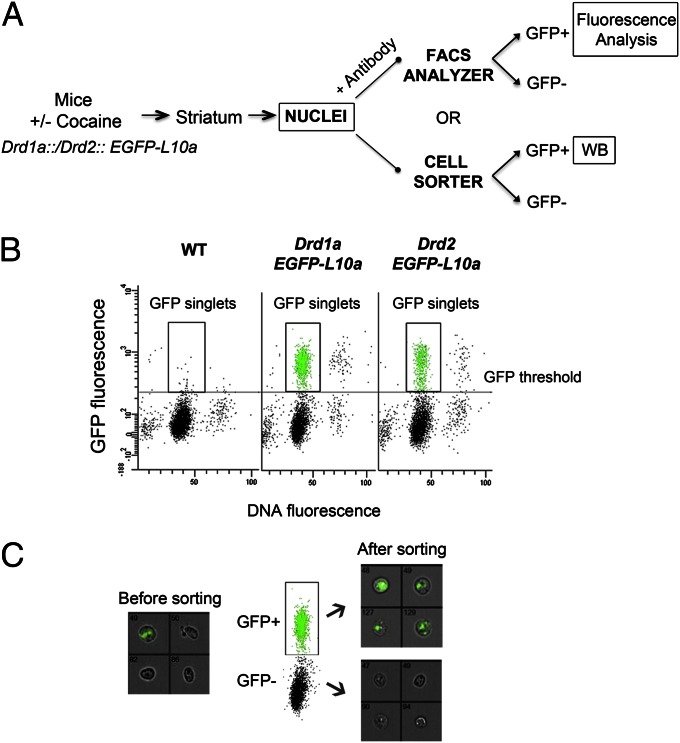
Fig. 1.
Fluorescence-activated sorting of nuclei from Drd1a::EGFP-L10a and Drd2::EGFP-L10a transgenic mice. (A) Outline of the procedure used to study histone PTMs in isolated neuronal nuclei populations. Histone PTM changes were either quantified using immunolabeling of permeabilized nuclei (+ antibody) and flow cytometry or using immunoblotting of sorted D1 and D2 nuclei (WB) (B) Typical flow cytometry dot plot and gating of D1 or D2 nuclei. Scatterplot of GFP fluorescence versus DyeCycle ruby (DCR) fluorescence of nuclei isolated from a WT mouse and Drd1a::EGFP-L10a or Drd2::EGFP-L10a transgenic mice. DCR binds quantitatively to the DNA and allows discrimination of singlets versus aggregated nuclei. The data from WT mice were used to set a threshold for the GFP signal background. (C) After sorting, nuclei are pure and intact as verified by Amnis ImageStream analysis. Representative results are for pre- and postsort fractions from Drd1a::EGFP-L10a mice. Bright-field and GFP channels are merged; original magnification 40×.