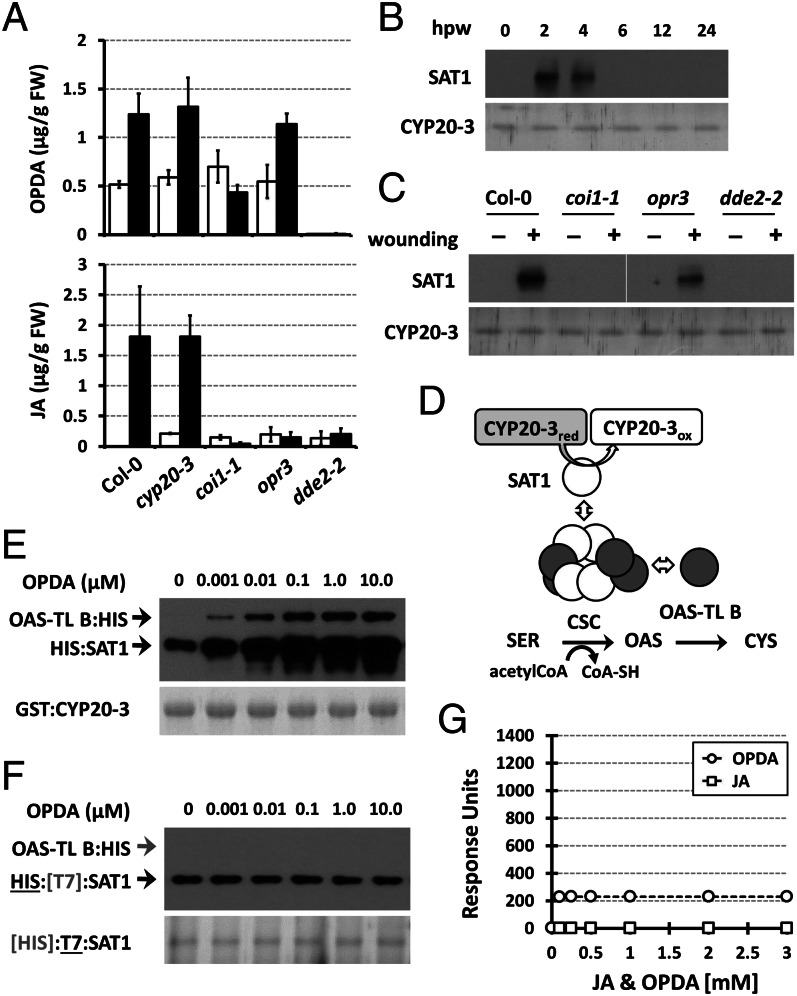
Fig. 3.
OPDA stimulates CYP20-3–SAT1 interaction and CSC formation. (A) Accumulation of OPDA and JA in wounded WT (Col-0) and mutant (cyp20-3, coi1-1, opr3, and dde2-2) plants. Jasmonates were extracted from leaves at 0 hpw (white bars) and 3 hpw (black bars; mean ± SD; n = 3). (B) Time-resolved in situ co-IP assays determining CYP20-3–SAT1 interaction in wounded WT (Col-0) plant. (C) In situ co-IP assays determining the CYP20-3-SAT1 interaction in WT (Col-0) and mutant (coi1-1, opr3, and dde2-2) plants at 3 hpw. (B and C) Total protein extracts were subjected to co-IP using anti-CYP20-3 antibody-coupled resin. (Lower) Silver-stained gels indicating amount of bait proteins (CYP20-3) used in each IP assay. Parallel immunoblots of proteins from co-IP with CYP20-3 were probed with polyclonal anti-SAT antibody (Upper). (D) Schematics of the plastid Cys biosynthetic pathway. (E and F) In vitro pull-down assays for determining the physical association of CYP20-3, SAT1, and OAS-TL B (E), and SAT and OAS-TL B (F), at various OPDA concentrations. GST:CYP20-3 fusion protein (E) or T7-tagged SAT1 (F) were used as a bait to pull down HIS:SAT1 and/or OAS-TL B:HIS proteins. (Lower) Coomassie blue-stained gels indicating amount of bait proteins used in each pull-down assay. Parallel immunoblots of proteins that copurified with the baits were probed with monoclonal anti-His antibody (Upper). (G) SPR analyses of SAT1 interactions with increasing concentrations of JA (□) and OPDA (○). The global fit of all data (n = 3) was integrated and plotted as concentration dependency by using ANEMONA.XLT (46).