Fig. 1.
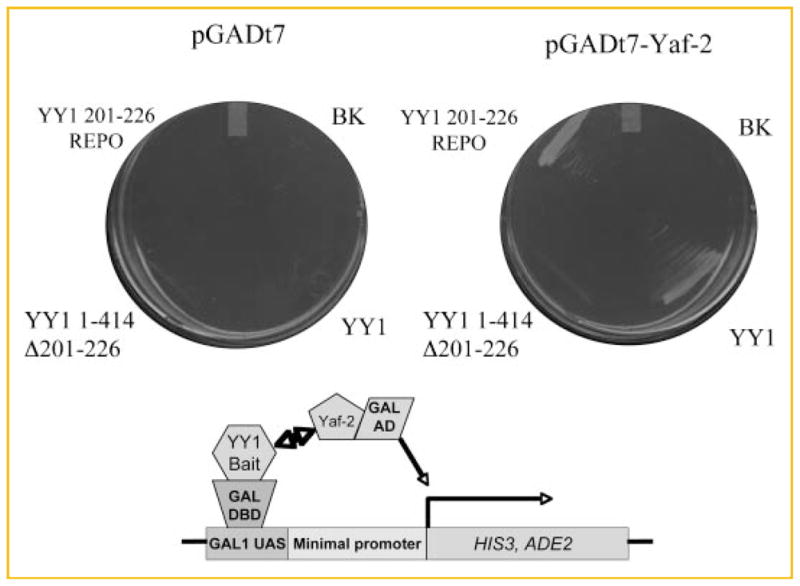
Yeast two hybrid detection of REPO–Yaf2 interactions. S. cerevisiae AH109 was transformed with the indicated bait and prey constructs. Cotransformants were passaged onto selective medium (Trp/Leu/Ade/His dropout medium). The plate on the left contains pGADt7 empty vector expressing the GAL4 activation domain (AD) and the plate on the right contains pGADt7 vector expressing the AD fused to full-length Yaf2 (residues 1–179, pGADt7-Yaf2). Bait constructs expressing GAL4 DBD fusions are as indicated: BK, pGBKt7 empty vector control; YY1, full-length YY1 1–414; YY1 1–414Δ201–226; YY1 201–226 REPO, YY1 residues 201–226. The scheme below diagrams the strategy used in the assay. Transcription of the nutritional markers HIS3 and ADE2 from interactions resulting from bait-prey binding interactions (two head arrow) is indicated.
