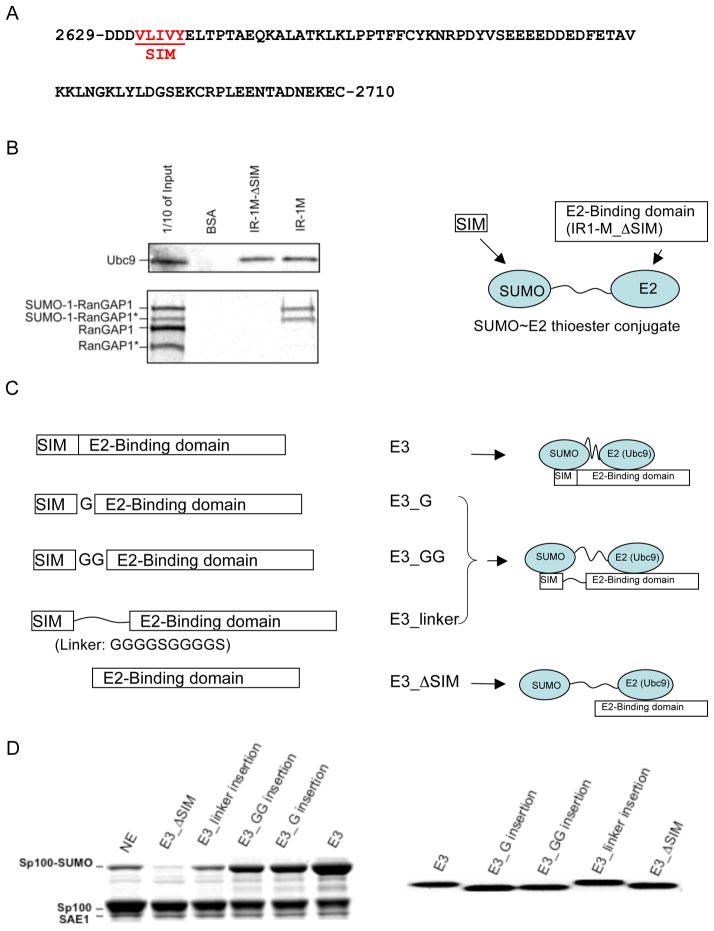
Figure 1.
Independence of the SUMO and Ubc9-binding sites in the IR1-M domain of RanBP2. (A) The IR1-M domain of RanBP2 is rich in Pro, polar and charged residues, similar to other disordered proteins. The SUMO-binding motif (SIM) is underlined and indicated in red. (B) The SUMO- and E2-binding regions in E3 are independent of each other. Pull-down experiments demonstrate that the SIM is required for binding SUMO (shown with SUMOylated RanGAP1). However, SIM deletion does not affect the interaction with Ubc9. Bound Ubc9 was detected by Western blot. RanGAP1 was labeled with 35S by in vitro transcription/translation reaction using rabbit reticulocyte extracts in the presence of 35S-methionine and was detected by autoradiography. Both RanGAP1 and SUMOylated RanGAP1 were produced during the translation process because rabbit reticulocyte extracts contained all the necessary components for SUMOylation of RanGAP1 (26). Asterisks indicate truncated version of RanGAP1 due to the presence of an ATG start codon in the middle of RanGAP1 cDNA. (C) Schematic diagrams of the E3 variants that contained linker insertions between the SIM and Ubc9-binding regions. Their expected effects on the SUMO~E2 thioester are shown to the right. (D) Comparison of ligase activity of the different E3 variants. The assay mixture (7 μL) containing 0.78 μM E1, 0.35 μM E2, 2.6 μM E3, 4 mM ATP, 22.7 μM SUMO, and 8.4 μM of Sp100 was incubated (37°C, 14 min) and quenched with freshly prepared DTT loading buffer (360 mM, 8.5 μL). The panel on the right shows Coomassie staining of the E3 variants to demonstrate that similar amounts were used in the activity assay.