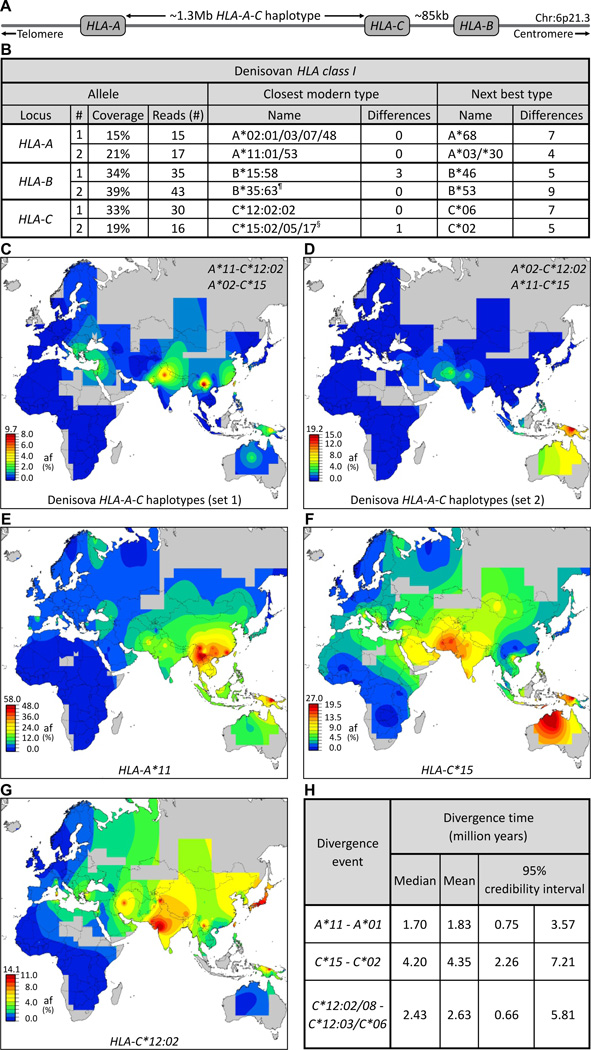
Fig.2.
Effect of adaptive introgression of Denisovan HLA class I alleles on modern Asian and Oceanian populations. (A) Simplified map of the HLA class I region showing the positions of the HLA-A, -B and -C genes. (B) Five of the six Denisovan HLA-A, -B and -C alleles are identical to modern counterparts. Shown at the left for each allele is the number of sequence reads (4) specific to that allele and their coverage of the ~3.5kb HLA class I gene. Center columns give the modern-human allele (HLA type) that has the lowest number of SNP mismatches to the Denisovan allele. The next most similar modern allele and the number of SNP differences are shown in the columns on the right. ¶, a recombinant allele with 5’ segments originating from B*40. §, the coding sequence is identical to C*15:05:02. (C–D) Show the worldwide distributions of the two possible Denisovan HLA-A-C haplotype combinations. Both are present in modern Asians and Oceanians but absent from Sub-Saharan Africans. (E–G) The distribution of three Denisovan alleles: HLA-A*11 (E), C*15 (F), and C*12:02 (G), in modern human populations shows they are common in Asians but absent or rare in Sub-Saharan Africans. (H) Estimation of divergence times shows that A*11, C*15 and C*12:02 were formed before the Out-of-Africa migration. Shown on the left are the alleles they diverged from, on the right are the divergence time estimates: median, mean, and range.