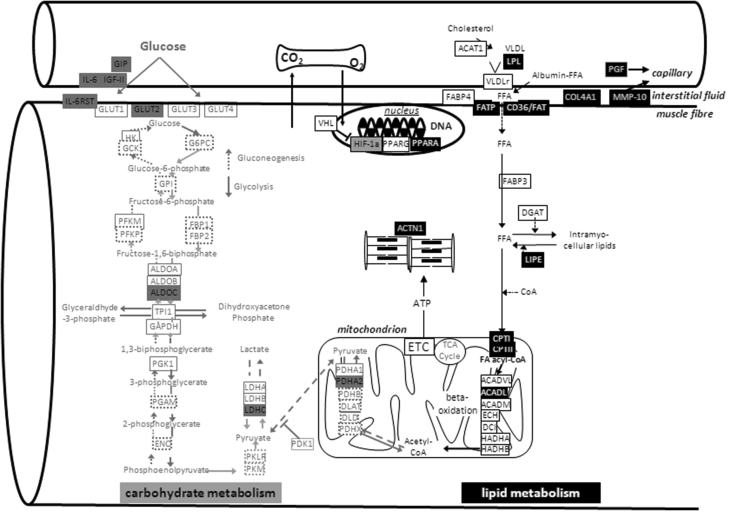
Fig. 4.
Summary of ACE I-allele affected pathways Genmapp visualizing the metabolic pathways holding gene transcripts with ACE I/D genotype dependent expression post exercise/training. The assessed transcripts are given in boxes with the colour coding indicting a significant genotype effect. ACADL long chain acyl-CoA dehydrogenase, ACTN1 actinin alpha 1, ALDOC aldolase C, COL4A1 collagen type IV alpha 1, CPTI carnitine palmitoyltransferase I, CPTII Carnitine palmitoyltransferase II, CD36/FAT cluster of Differentiation 36)/fatty acid translocase, ETC. electron transport chain, FATP fatty acid transport protein, GIP glucose-dependent insulinotropic peptide, Glut2 Glucose transporter 2, HIF-1a subunit alpha of hypoxia-inducible factor 1, HO-1 heme oxygenase 1, IGF-II insulin-like growth factor II, IL-6 interleukin 6, Il-6RST interleukin 6 receptor signal transducer, LDHC lactate dehydrogenase C, LIPE hormone sensitive lipase transcript, LPL lipoprotein lipase, MMP-10 metalloproteinase 10, PDHA2 pyruvate dehydrogenase alpha 2, PGF placental growth factor, PPARA Peroxisome proliferator-activated receptor alpha, VHL von Hippel Landau tumour suppressor. For further abbreviations consult www.expasy.org