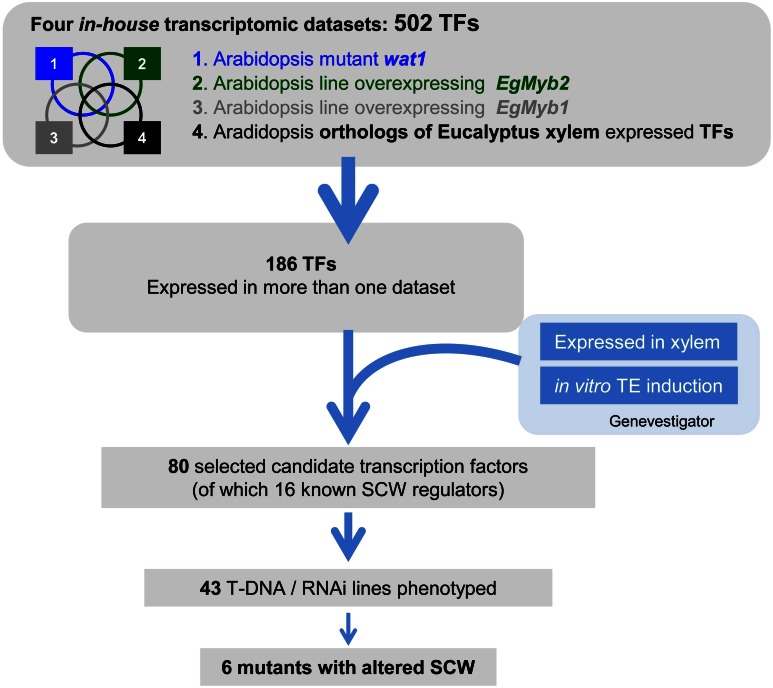
Figure 1.
Overall strategy to identify transcription factors (TFs) involved in SCW formation. Four in-house SCW formation-related transcriptomic datasets were crossed to select TFs present in more than one dataset/experimental condition. These TFs were further screened against publicly available large-scale transcriptomic datasets, to select those highly or preferentially expressed in the organs and/or tissues of interest. This led to a list of 80 candidate genes, of which we phenotyped the 42 T-DNA insertion mutants and/or RNAi transgenic plants available. The four in house SCW related transcriptomic datasets include (1) the fiber SCW-deficient wat1 Arabidopsis mutant, (2) Arabidopsis lines over-expressing the SCW master activator EgMYB2, (3) Arabidopsis lines over-expressing the SCW master repressor EgMYB1, and (4) Arabidopsis orthologs of Eucalyptus xylogenesis-related genes.