Figure 5.
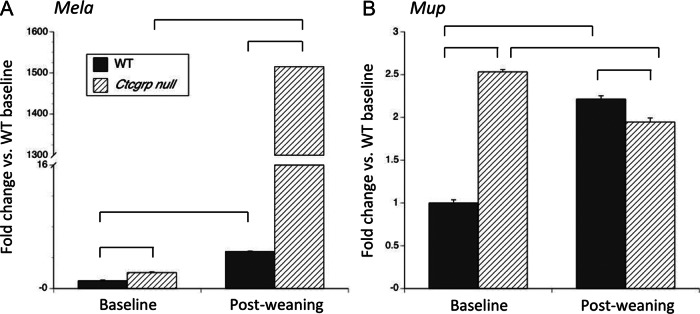
qPCR of Mela and Mup at day 7 postweaning vs baseline. The largest differential expression on the microarray occurred in Mela and Mup, neither of which has previously been shown to be expressed in bone or marrow. In A, qPCR confirmed the microarray findings of up-regulation of Mela during postweaning in both genotypes, with the greater increase achieved in Ctcgrp null mice. On the microarray, each of the 19 different Mup genes had 1.9-fold increased expression in Ctcgrp null mice during postweaning as compared with baseline (Supplemental Table 3) or 5-fold higher expression in Ctcgrp null mice during postweaning as compared with WT baseline (Supplemental Table 4). Shown in B is the result of a single qPCR primer set common to all Mup genes, which suggests that Mup gene expression was 2.5-fold higher in Ctcgrp null mice than WT mice at baseline, whereas during postweaning it decreased slightly in Ctgrp null mice but increased 2-fold in WT mice. It is unknown whether Mela or Mup plays any role in regulating skeletal metabolism during postweaning. Braces indicate P < .01.
