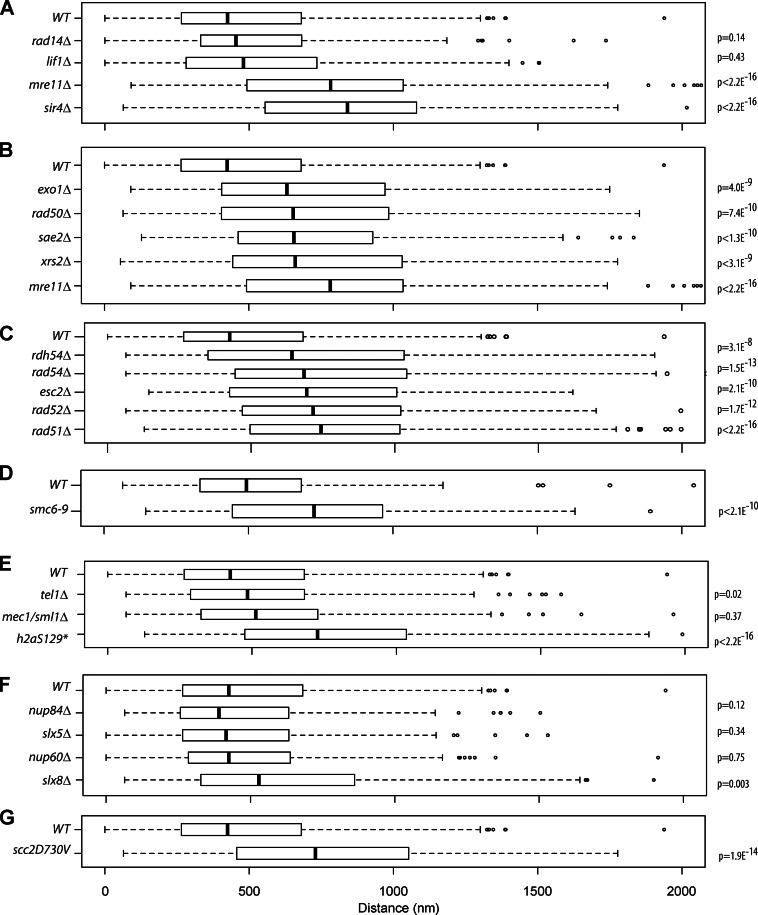
Figure 4.
DNA DSB repair proteins contribute to HM long-range interactions. (A) Boxplots of the distance between TetR-YFP and CFP-LacI foci in a given strain. Strains containing deletions in rad14Δ (n = 186), lif1Δ (n = 143), mre11Δ (n = 443), and sir4Δ (n = 134). (B) Strains containing deletions in early HR repair proteins (exo1Δ, n = 214; rad50Δ, n = 414; sae2Δ, n = 161; xrs2Δ, n = 123; and mre11Δ, the mre11Δ data are the same as in A and are simply shown for easy comparison). (C) Strains containing deletions in downstream HR repair proteins (rdh54Δ, n = 215; rad54Δ, n = 223; esc2Δ, n = 167; rad52Δ, n = 163; and rad51Δ, n = 676). (D) WT and a temperature-sensitive smc6-9 strain after 2 h at 37°C. (E) The checkpoint proteins tel1Δ and mec1Δ/sml1Δ and a H2A mutant (tel1Δ, n = 143; mec1Δ/sml1Δ, n = 184; and hta1-129*Δ hta2-129*Δ, n = 437). (F) Nuclear pore proteins and ubiquitin pathway proteins (nup84Δ, n = 215; slx5Δ, n = 196; slx8Δ, n = 215; and nup60Δ, n = 210). (G) A point mutant scc2D730V. The WT data in all panels (except D) are the same as in A and are simply shown for easy comparison. The boxes represent the middle 50% of data points with the black lines showing the median of distances. Outliers are defined as distances >1.5 times the interquartile range (dashed lines) and are represented by open circles.