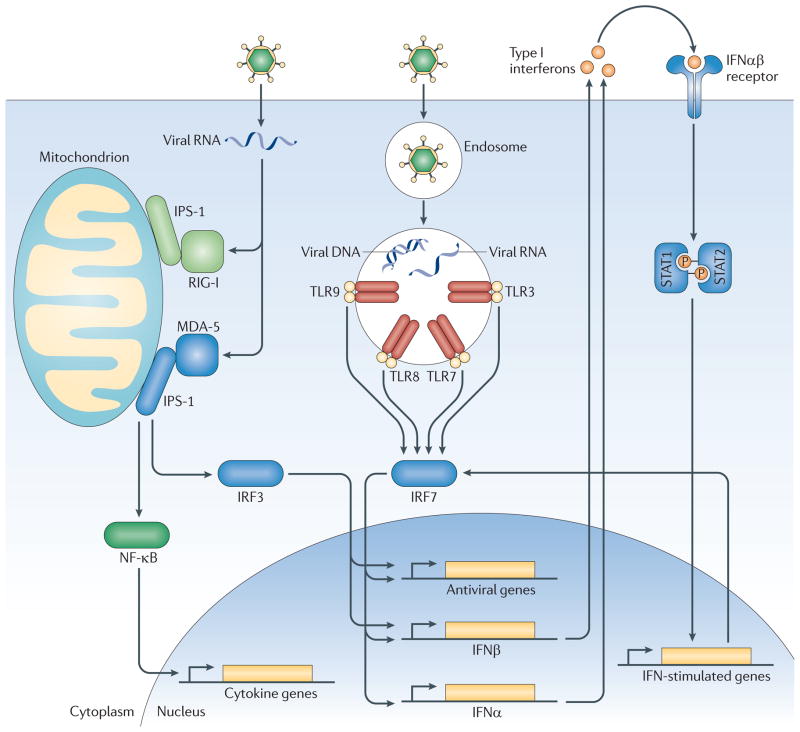
Figure 1. Components of the anti-viral immune response genetically linked to Type I Diabetes.
Viruses are recognized in general by two separate pathways. Intrinsic recognition occurs through detection of viral nucleic acids by cytosolic RLRs and other nucleic acid sensors in the infected cells. MDA-5/IFI1H is a cytosolic RLR that can recognize Picornavirus genomes. Activated MDA-5 activates the transcription factors IRF-3 and NFκB. In contrast, extrinsic recognition of virus occurs through Toll-like receptors (TLR) 3, 7, 8 and 9, which can recognize viral DNA and RNA within endosomes and activate transcription factors IRF-7 and NF-κb. IRF-3 and IRF-7 function in homo- or heterodimers to initiate transcription of Type I interferons and other anti-viral genes, as shown. Via NF-κB, both pathways induce the expression of pro-inflammatory cytokines. Secreted Type I interferons bind to the IFNαβR on the cell surface, which signals via STAT1 and STAT2 to induce expression of ~300 interferon-stimulated genes (ISGs) including MDA-5 and IRF-7. Type 1 diabetes is linked to genetic polymorphisms in MDA-5/IFI1H, IRF-7, and an IRF-7 driven network of 305 genes (likely the depicted network of IFN response genes.) Interestingly, T1D is predicted to correlate with robust responses through these pathways. For MDA-5, polymorphisms that result greater expression correlate with disease, whereas rare alleles that result in loss of or reduced function are protective. Similarly, T1D-associated polymorphisms in the genomic locus regulating the IRF-7 driven network predict increased expression of this network in T1D.