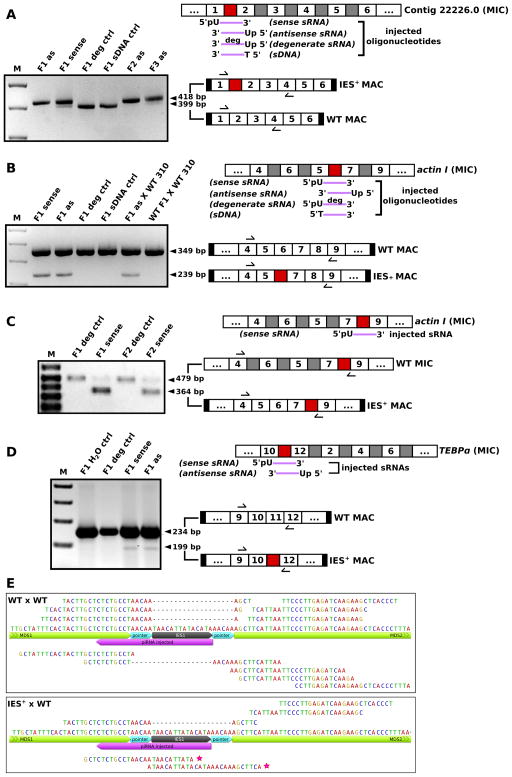
Figure 5. Small RNA injection leads to heritable retention of normally-deleted genomic regions.
PCR amplification of total DNA from exconjugant cells after corresponding synthetic sRNA injection reveals programmed IES retention (labeled IES+ MAC). (A) PCR assay for the presence of a larger IES+ MAC PCR product if the targeted conventional IES between MDS1–2 is retained in MAC Contig22226.0. Bands corresponding to wild-type (WT) MAC PCR products are also indicated. (B) PCR assays for a smaller actin I MAC PCR product if the scrambled IES between MDS5–7 is retained instead of being replaced by the longer, scrambled MDS6. (C) Selective PCR assay for the presence of a smaller actin I MAC PCR product if the targeted IES between MDS7–9 is retained; larger bands correspond to the scrambled precursor MIC DNA. (D) PCR assay for the presence of a smaller actin I MAC PCR product if the scrambled IES between MDS10–12 is retained. MIC contig structure, synthetic oligonucleotides injected (purple), and PCR products are shown schematically on the right. MDSs (white boxes with numbers) and IESs (grey and red boxes) are not drawn to scale. Red boxes represent IESs that are targeted by injected RNAs, and black boxes denote short telomeres on nanochromosomes. Arrows schematically indicate primer positions. (E) Exconjugant cells from a backcross between the Contig22226.0 IES1+ strain and a parental wild-type strain (IES+× WT) produce new IES-containing piRNAs, but wild-type mating cells (WT × WT) do not. 25–28 nt 5′-U reads are plotted. Forward and reverse mappings are displayed above and below the relevant portion of the full-length annotated contig, respectively. Stars indicate two new IES-containing piRNAs. Gaps indicate WT reads mapping across the MDS6–7 junction. See also Figure S6 and sequence alignments of all PCR products in a supplemental file.