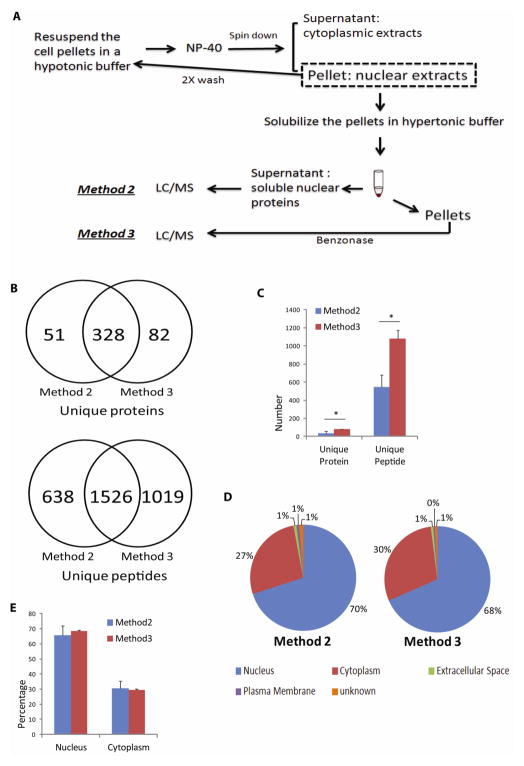
Fig. 2. Effect of nuclease treatment on nuclear protein recovery.
(A) After initial isolation of the nuclear pellets from SHSY-5Y cells that included repeated washes to remove loosely associated cytoplasmic proteins, nuclear proteins were extracted with the hypertonic buffer, and the supernatants were digested either immediately (see Materials and Methods: Method 2) or after Benzonase® nuclease digestion of the pellets (see Materials and Methods: Method 3). Free nuclear proteins from Method 2 and DNA-associated proteins recovered from Method 3 were identified by LC/MS/MS analysis. (B) and (C) Comparison of the unique proteins and peptides identified from either method. (B) shows data from one of the representative experiment. In addition to a large degree of protein and peptide overlap found between the two methods, nuclease digestion of DNA resulted in the identification of ~40% more peptides and 20% more proteins in Method 3, when compared with Method 2 alone (* P≤ 0.05). Method 2 (Blue bar); Method 3 (Red bar). (D) and (E) Comparison of IPA-annotated nuclear proteins between the two methods. (D) shows data from one of the representative experiment. Relative percentages of nuclear proteins between the two methods were comparable. Subcellular localization annotation: Nucleus (Blue); Cytoplasm (Red); Extracellular Space (Green); Plasma Membrane (Purple); Unknown (Orange).