Abstract
Somatic gonadal niche cells control the survival, differentiation, and proliferation of germline stem cells. The establishment of this niche-stem cell relationship is critical, and yet the precursors to these two cell types are often born at a distance from one another. The simple C. elegans gonadal primordium, which contains two somatic gonad precursors (SGPs) and two primordial germ cells (PGCs), provides an accessible model for determining how stem cell and niche cell precursors first assemble during development. To visualize the morphogenetic events that lead to formation of the gonadal primordium, we generated transgenic strains to label the cell membranes of the SGPs and PGCs and captured time-lapse movies as the gonadal primordium formed. We identify three distinct phases of SGP behavior: posterior migration along the endoderm towards the PGCs, extension of a single long projection around the adjacent PGC, and a dramatic wrapping over the PGC surfaces. We show that the endoderm and PGCs are dispensable for SGP posterior migration and initiation of projections. However, both tissues are required for the final positioning of the SGPs and the morphology of their projections, and PGCs are absolutely required for SGP wrapping behaviors. Finally, we demonstrate that the basement membrane component laminin, which localizes adjacent to the developing gonadal primordium, is required to prevent the SGPs from over-extending past the PGCs. Our findings provide a foundation for understanding the cellular and molecular regulation of the establishment of a niche-stem cell relationship.
Keywords: somatic gonad precursor, primordial germ cell, niche, gonad, laminin, cell migration
INTRODUCTION
Through their ability to self-renew and differentiate, stem cells are responsible for the formation and homeostasis of many diverse cell types and tissues. Many stem cells have been shown to reside in a specialized microenvironment called the niche, where supporting cells provide signals that control stem cell proliferation, differentiation, or survival (reviewed by Hubbard, 2007; Byrd and Kimble, 2009; Losick et al., 2011; Lehmann, 2012); in the absence of these signals from the niche, stem cells can divide inappropriately or die, leading to abnormal growth or tissue loss. An important yet poorly understood question is how niche cell precursors and stem cell precursors first come together during development to establish a functional niche–stem cell interaction. Given the small number of model systems where the identities of both stem and niche precursor cells are known with certainty, this question has been challenging to address (reviewed by Morrison and Spradling, 2008).
One clear and conserved example of a niche–stem cell interaction is that between somatic gonad niche cells and germline stem cells. Interactions between these two cell types have been studied extensively in the larval and adult C. elegans gonad. The hermaphrodite gonad is comprised of two equivalent arms in which germline stem cells proliferate at the distal ends and differentiate into gametes more proximally (reviewed by Hubbard, 2007; Byrd and Kimble, 2009). A somatic cell called the distal tip cell (DTC) wraps around the distal end of each gonad arm and signals through the Notch pathway to control the proliferation and differentiation of the germline stem cells. By tracing the lineages of gonadal cells, it has been possible to establish that all the somatic cells of the adult gonad (including the DTCs) arise from just two somatic gonad precursor cells (SGPs), and the entire germ line arises from two primordial germ cells (PGCs) (Kimble and Hirsh, 1979; Sulston et al., 1983). The two SGPs and two PGCs are born during embryogenesis and undergo coordinated divisions during larval stages to produce the entire gonad and germ line.
The establishment of the interaction between the two cell types occurs during the middle of embryogenesis, when the two SGPs and two PGCs first come together to form the gonadal primordium (Sulston et al., 1983; reviewed by Hubbard and Greenstein, 2000). Classic laser ablation experiments showed that essential interactions between the SGPs and PGCs occur even at these early stages. For example, if the SGP precursors are ablated prior to gonadal primordium assembly, the PGCs die (Kimble and White, 1981). And if the SGPs are ablated after the gonadal primordium forms, PGCs survive but cannot proliferate (Kimble and White, 1981). Therefore, even within the gonadal primordium, the SGPs function as a niche for PGCs, controlling their survival and proliferation. Despite the importance of the interaction between SGPs and PGCs, relatively little is known about how these two cell types first come together to form a functional niche-stem cell interaction.
The SGPs and PGCs are born in different regions of the embryo, and both cell groups undergo morphogenetic movements to form the gonadal primordium. The two PGCs (called Z2 and Z3) are born on the posterior ventral surface of the embryo (Sulston et al., 1983) and adhere tightly to adjacent endoderm cells. Morphogenetic movements of the endoderm pull the PGCs from the surface of the embryo into the center where the gonadal primordium will form (Chihara and Nance, 2012). By contrast, the two SGPs (called Z1 and Z4) are born in more anterior regions within the interior of the embryo and must then migrate posteriorly to join the PGCs (Sulston et al., 1983). Cell migration is a conserved feature of gonadal primordium development in many species, although it is the PGCs that migrate long distances to join the SGPs in vertebrates such as zebrafish and mouse, and PGCs and SGPs are both known to migrate in Drosophila (reviewed by Richardson and Lehmann, 2010). However, the mechanisms used to guide C. elegans SGPs towards the PGCs and to stop once finding them are unknown.
Here, as an initial step towards understanding how interactions between somatic gonadal niche cells and germline stem cells are first established, we have analyzed C. elegans SGP migration and gonadal primordium assembly in vivo using fluorescence time-lapse microscopy. We describe three distinct phases of SGP behavior: posterior migration along the endoderm, extension of projections around adjacent PGCs, and wrapping over the PGC surfaces. We find that neither endoderm nor PGCs are required for SGP migration or initiation of projections, but we identify distinct contributions for these cells in the positioning of SGPs at the end of their migration, in SGP projection morphology, and in promoting wrapping behavior around the PGCs. Finally, we demonstrate that the basement membrane component laminin is required to ensure that SGPs do not over-extend past the PGCs. Our findings provide a foundation for identifying the cellular and molecular cues that promote the assembly of a niche-stem cell interaction during development.
MATERIALS and METHODS
Strains
The following strains were used: N2 (wild type); FT853: zuIs60 [pie-1P::secretedGFP]; zuIs244 [nmy-2P::PGL-1::RFP]; ltIs44 [pie-1P::mCherry-PHPLC1∂1] (Chihara and Nance, 2012); FT854: xnIs307 [hnd-1P::GFP-PHPLC1∂1]; unc-119(ed3); xnIs91 [end-1P::mCherry-PHPLC1∂1]; FT885: xnIs307; xnIs91; zuIs244; FT938: end-1 end-3; irEx568 [end-1(+), end-3(+), sur-5::RFP]; xnIs307; xnIs91; zuIs244; FT968: hmr-1(zu389); xnIs307; xnIs91; zuIs244; FT977: xnIs307; unc-119(ed3); ltIs44; FT983: xnIs307; unc-119(ed3); xnIs360 [mex-5P::mCherry-PHPLC1∂1::nos-2UTR]; IM253: urEx131[IM#px41.1 (lam-1::gfp), pRF4 (rol-6)] (Kao et al., 2006); SS149: mes-1(bn7) (Strome et al., 1995). Some strains were provided by the CGC.
Transgene construction and worm transformation
hnd-1P::GFP-PHPLC1∂1 and mex-5P::mCherry-PHPLC1∂1::nos-2UTR were created using Multisite Gateway™ (Invitrogen), the pCFJ150 destination vector (Frokjaer-Jensen et al., 2008), and the following entry clones: 5′ entry clones: pMRR2 (hnd-1P) (this study) and pJA252 (mex-5P) (Zeiser et al., 2011), middle entry clones pJN535 (GFP-PHPLC1∂1) (this study) and pMRR7 (mCherry-PHPLC1∂1) (this study), and 3′ entry clones pCM1.36 (tbb-2UTR) (Merritt et al., 2008) and pDC10 (nos-2UTR) (Chihara and Nance, 2012). To make pJN535 (GFP-PHPLC1∂1), GFP fused to the rat PLC1∂1 PH domain (Audhya et al., 2005) was PCR amplified and recombined into pDONR221 (Invitrogen) using Gateway™.
To make pMRR2 (hnd-1P), the first two exons of hnd-1 plus 1587bp upstream of its start ATG, a sequence identified by Mathies et al. (2003) as promoting SGP expression, was PCR amplified and cloned into pDONR-P4-P1R (Invitrogen) using Gateway™. Primers:
oMRR1: GGGGACAACTTTGTATAGAAAAGTTGTTAGGTGCCGAGTAGCATATGAC
oMRR3: GGGGACTGCTTTTTTGTACAAACTTGCATCTCATTTCCTCCCAGCAATT
To make pMRR7 (mCherry-PHPLC1∂1), mCherry was PCR amplified from pJA281 (mex-5P::mCherry), and used to replace GFP in middle entry clone pJN535 (GFP-PHPLC1∂1).
Integrated transgenic lines were created by microparticle bombardment of unc-119(ed3) as described (Praitis et al., 2001).
Microscopy
All images were of embryos mounted on 4% agarose at the 1-2 cell stage and allowed to develop to the appropriate stage at 25°C. 4D DIC movies were acquired using a Zeiss AxioImager, 63× 1.4NA objective, Nomarski optics, an Axiocam MRM camera and AxioVision software. Z-stacks at 0.5μm intervals were captured every 45 seconds, and SGPs, PGCs, and various endoderm cells were identified by lineaging with the help of MTrackJ software (Meijering et al., 2012). SGP birth location was measured from DIC movies of wild type, mes-1(bn7) mutant, and end-1 end-3 mutant embryos. Combined fluorescence and DIC Z-stacks (1–2μm intervals) were also acquired using this microscope.
4D fluorescence movies were acquired using a Leica SP5 confocal microscope, 63x 1.3NA water-immersion objective, 488 and 594 nm laser. Embryos for transverse movies were mounted as previously described (Rasmussen et al., 2012). All movies and Z-stacks were analyzed and quantified in ImageJ. Images were cropped, rotated, and levels were adjusted in Adobe Photoshop. Supplemental movies were created using ImageJ or Imaris.
Laser irradiation
Laser irradiation was performed as previously described (Chihara and Nance, 2012), except MSap or MSpp were targeted, and after irradiation, embryos were allowed to develop to the desired stage. We did not observe a difference in the results of MSap (n = 10) and MSpp (n = 17) ablations. Irradiated embryos were analyzed only if the expected SGP was missing (as detected by both hnd-1P::GFP-PHPLC1∂1 and DIC imaging), and non-targeted lineages were normal.
RNAi
RNAi was performed by the feeding method as previously described (Chihara and Nance, 2012) using empty vector pPD129.36 in HT115 as a negative control. mes-1 RNAi was performed at 25° and lam-1 and lam-2 RNAi was performed at 20° or 25°.
RNAi feeding constructs targeting lam-1 and lam-2 were cloned into the pPD129.36 RNAi feeding vector (Timmons and Fire, 1998) digested with EcoRV. lam-1 and lam-2 exon-rich DNA was amplified from genomic DNA with the following primers:
lam-1: GTGCCGACATTACTCATTACG, CTCCGAGTCTTGGATCTC
lam-2: CCCAAGAATCAATGAACTCGAA, CATCCATTGGCACTGAATCC
40bp homology arms to sequences flanking the EcoRV sites in pPD129.36 were included at the 5′ end of each primer. Inserts and vector were combined using Gibson end-joining (Gibson et al., 2009).
Immunostaining
Embryos were fixed, stained, and imaged as described (Anderson et al, 2008). The following antibodies were used: rabbit α-GFP 1:1000 (Abcam), rat α-mCherry 1:10 (Rottach et al., 2008), and chicken α-LAM-3 (Huang et al., 2003).
RESULTS
Somatic gonad precursors migrate posteriorly and wrap around the PGCs
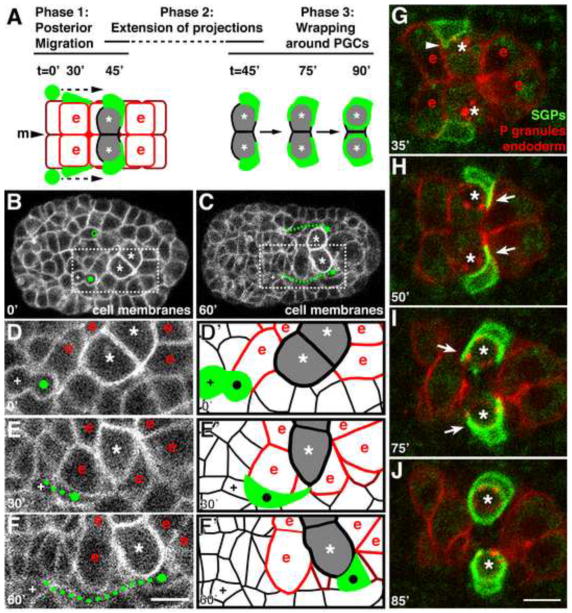
To observe how the SGPs migrate to the PGCs and determine the path that they follow, we recorded three-dimensional fluorescence time-lapse (‘4D’) movies of embryos expressing pie-1P::mCherry-PHPLC1∂1, which labels the plasma membrane of all cells (Fig. 1B–F; D′–F′ are schematics of D–F). The two SGPs (green in Fig. 1A, marked with circles in Fig. 1B,D) arise from separate lineages but are born at a similar time (~250 minutes after the first cleavage; Sulston et al., 1983), one on each side of the midline and lateral to the anterior-most cells of the endoderm (Fig. 1A,B,D; Fig. 4D). Their birth position corresponds to a location of 48±2% embryo length (n = 8 embryos; values are averages ± standard deviation here and throughout the paper), as measured from the anterior tip (0%) to posterior pole (100%) of the embryo (a distance of ~50μm). At this stage, the two PGCs (asterisks, Fig. 1B,D; Fig. 4D) lie more posteriorly, on opposite sides of the midline, adjacent to one another in a pocket formed by the endoderm cells. After their birth (circles, Fig. 1B,D), the SGPs immediately began to elongate in shape (compare Fig. 1D,E), then migrate posteriorly just lateral to the endoderm (Fig. 1E,F). At 27±9 minutes after their birth, each SGP first contacted its ipsilateral PGC (Z1 adjacent to Z2 on the right side of the embryo; Z4 adjacent to Z3 on the left; Fig. 1E). At this point, the SGPs stopped elongating further, but continued moving posteriorly until they came to lie just posterior and lateral to the PGCs and just anterior and lateral to the posterior-most ventral endodermal cells (row 8; Leung et al., 1999) (70±3% embryo length, n = 23 embryos; Fig. 1F). SGP migration lasted 60±6 minutes and covered an average distance of 12μm (Fig. 1C, Fig. 4D; n = 7 embryos). By contrast, the sister cell of each SGP (marked by a +, Fig. 1B–F) remained in approximately the same location over this time period, demonstrating that the posterior movement of the SGPs was active migration rather than the result of tissue movements. In summary, the SGPs begin to migrate posteriorly immediately after their birth, appear to be in close association with the endoderm throughout their migration, and stop migrating when they reach a location just posterior to the PGCs (Fig. 1A).
Figure 1. SGPs migrate posteriorly, extend projections, and wrap around the PGCs.
In this and subsequent figures, time shown is minutes after SGP birth. Anterior is to the left; ventral view. Embryo is ~50μm in length. (A) Schematic of the three phases of SGP morphogenetic behaviors during gonadal primordium assembly. SGPs are green, ventral endodermal cells are outlined in red and labeled (‘e’), dorsal endodermal cells are outlined in maroon. PGCs are shaded in gray and indicated with an asterisk. “m” marks the midline. (B,C) Stills from a 4D confocal movie of embryo expressing the cell surface membrane marker pie-1P::mCherry-PHPLC1∂1. PGCs are marked with an asterisk, SGPs are indicated with a filled green circle, and SGP migratory path indicated with a dashed green line. The SGP sister is marked with ‘+’, and open green circle in (B) is the parent of the SGP that has not yet divided. (D–F) Blow-up of boxed region shown in (B,C) at indicated times, labeled as above; ‘e’ cells are endoderm adjacent the PGCs. (D′–F′) Schematic of panels E–F showing cell outlines and position of SGPs; cells are marked as in panel A. (G–J) Stills from a 4D confocal movie of embryo expressing hnd-1P::GFP-PHPLC1∂1 (SGPs), end-1P::mCherry-PHPLC1∂1 (endoderm), and nmy-2P::PGL-1::RFP (PGC-specific P-granules). PGCs are indicated with asterisks. (G) The SGPs extend small protrusions over the endoderm (arrowhead), (H) then extend long projections posterior to the PGCs (arrow). (I) The SGPs subsequently expand over to cover the ventral and anterior surfaces (arrow), (J) and finally wrap completely around the PGCs. Scale bars are 5μm.
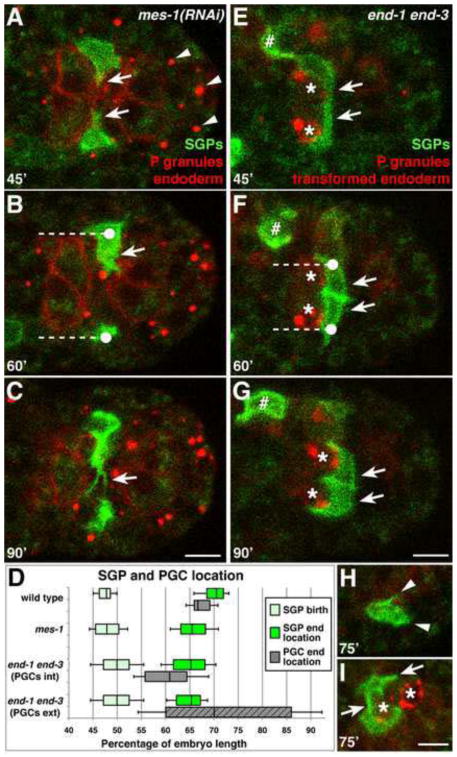
Figure 4. PGCs are required for SGP wrapping, and PGCs and endoderm are required for SGP positioning and projection morphology.
Stills from a 4D confocal movie of embryos of the indicated genotype expressing hnd-1P::GFP-PHPLC1∂1 (SGPs), end-1P::mCherry-PHPLC1∂1 (endoderm), and nmy-2P::PGL-1::RFP (PGC-specific P-granules). PGCs are indicated with asterisks. See Fig. 1G-J for comparison to wild type. (A–C) Arrowheads mark P-granules in transformed PGCs. (A) In mes-1(RNAi) embryos, SGPs extend thicker projections (arrows in A,B), (B) and do not migrate quite as far posteriorly (dashed line indicates distance migrated from average SGP birth location). (C) SGPs continue to extend projections and fail to transition to wrapping behaviors. (D) Quantification of SGP and PGC position in indicated genotypes. Data for end-1 end-3 embryos is divided into those with internal PGCs “int” (solid gray rectangle) and external PGCs “ext” (hatched gray rectangle). The median (line in box), 25th and 75th percentiles (box boundaries), and standard deviation (error bars) are shown. The SGP birth locations were not significantly different between the four categories. The difference between SGP birth and end location was significant for each category. The difference between wild-type SGP end location and the end locations for the other three genotypes was also significant (p<0.001, two-tailed Student’s t-test). (E–H) (E) In end-1 end-3 embryos, the SGPs extend shorter, thicker projections (arrows), (F) and do not migrate as far posteriorly (dashed line indicates distance migrated from average SGP birth location). SGPs can shift more medially and come in contact with each other (arrows in F,G). (G) SGPs eventually wrap around PGCs. Expression of hnd-1P::GFP-PHPLC1∂1 in a misplaced non-SGP cell is indicated (#). (H) In end-1 end-3 embryos with PGCs external, SGPs extend short projections (arrowheads) but fail to wrap around any cell. (I) end-1 end-3 embryo in which one SGP contacts both PGCs, and one PGC is briefly wrapped by both SGPs (arrows). Scale bars are 5μm.
To more easily visualize the dynamic movements of the migrating SGPs and the cells that they contact, we constructed a transgene that labels the surfaces of the SGPs with green fluorescent protein (hnd-1P::GFP-PHPLC1∂1), and combined it with transgenes that label the surfaces of the endoderm with mCherry (end-1P::mCherry-PHPLC1∂1) and the PGCs with the P granule marker nmy-2P::PGL-1::RFP. hnd-1P::GFP-PHPLC1∂1 was first visibly expressed in SGPs when they had migrated about half of the distance to the PGCs, and expression peaked after the two cell types met (Fig. 1G–J). In multi-color 4D movies, we observed the migrating SGPs extending short protrusions both posteriorly and medially over the surface of the endoderm (arrowhead, Fig. 1G; Movie S1). Shortly after the SGPs came into contact with the PGCs, they each extended a single long projection medially, around the posterior edge of the PGCs and just anterior to the endoderm row 8 cells (45±9 minutes after their birth; arrows in Fig. 1H; n = 7 embryos). While the projections from Z1 and Z4 could touch briefly, in general each SGP respected the midline: Z1 remained on the right side of the embryo and extended around Z2, while Z4 remained on the left side and extended around Z3 (Fig. 1G–J). Finally, ~90 minutes after the SGPs were born, they each finished wrapping around the adjacent PGC (Fig. 1A,I,J).
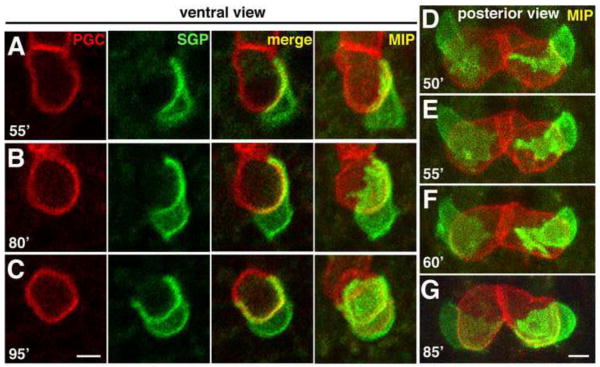
To determine whether the SGP projections wrapped directly around the PGC surfaces, we constructed a transgene that labels the PGC surfaces with mCherry (mex-5P::mCherry-PHPLC1∂1::nos-2UTR) and combined it with the SGP surface marker. Multi-color 4D movies showed that the SGP projections were indeed tightly associated with the surfaces of the PGCs as soon as the two cell types initiated contact with each other (Fig. 2A–C; Movie S2; n = 9 embryos). In movies of embryos imaged transversely from their posterior end, we observed that these projections were dynamic lamellar structures that first extended towards the midline (where the PGCs contacted one another) then broadened to cover the posterior surface of the PGCs (Fig. 2D–F; n = 8 embryos). The SGP projections continued to expand over the ventral surfaces of the PGCs and around to their anterior sides (arrows in Fig. 1I, 2B–C), ensheathing the ventral surfaces of the PGCs and forming the four-celled gonadal primordium [Fig. 2G; the dorsal PGC surfaces protrude into the endoderm (data not shown and Sulston et al., 1983)]. All together, our time-lapse imaging experiments revealed three distinct phases of SGP behavior leading to gonadal primordium assembly (summarized in Fig. 1A). In Phase 1, the SGPs migrate posteriorly along the endoderm. In Phase 2, the SGPs extend a single, long projection around the adjacent PGC. Finally, in Phase 3, the SGPs dramatically wrap around the PGCs, ensheathing their ventral surfaces.
Figure 2. SGP wrapping over PGC surfaces.
(A–C) Stills from 4D confocal movie of embryo expressing hnd-1P::GFP-PHPLC1∂1 (SGPs) and mex-5P::mCherry-PHPLC1∂1::nos-2UTR (PGCs) imaged from a ventral perspective; a central focal plane (left three panels) as well as a maximum intensity (MIP) projection all the way through the two cells is shown. (D–G) Same as A–C except imaged transversely from a posterior perspective, maximum intensity projection shown. In both sets of images, the SGPs can be seen extending projections over the posterior surfaces of the PGCs (A,D–F), then expanding and wrapping over the ventral surfaces of the PGCs (B,C). Scale bars are 2.5μm.
SGPs migrate, extend projections, and wrap the PGCs independently of each other
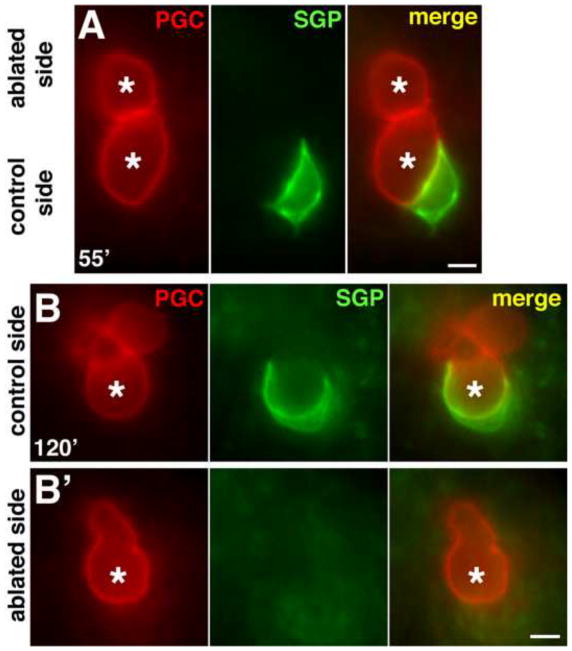
Although Z1 and Z4 are born and migrate posteriorly on opposite sides of the embryo, they move through the three phases of SGP behavior in unison and with mirror-symmetry across the midline. These observations raised the possibility that either the SGPs communicate with one another to coordinate their morphogenesis, or alternatively, that the SGPs are each influenced by the same global signals. To distinguish between these two possibilities, we killed the ancestor of one SGP by laser-irradiation and observed the behavior of the remaining SGP using transgenes to mark the SGP and PGC surfaces. We found that the remaining SGP always migrated to the normal location (70±3% embryo length; n = 7 embryos; Fig. 3A), extended a long projection around the posterior side of the ipsilateral PGC (7/7 embryos at t=60′; Fig. 3A), and eventually wrapped entirely around that PGC (20/20 embryos at t=120′; Fig. 3B). In laser-irradiated embryos, we never observed the single SGP significantly over- or under-migrate, or wrap around the contralateral PGC. We conclude that even though the two SGPs undergo mirror-symmetric and synchronous morphogenetic movements, Z1 and Z4 are not dependent on each other for their posterior migration, extension of projections, or wrapping behaviors.
Figure 3. SGPs migrate, extend projections, and wrap PGCs independently of each other.
(A–B′) Live embryos expressing hnd-1P::GFP-PHPLC1∂1 (SGPs) and mex-5P::mCherry-PHPLC1∂1::nos-2UTR (PGCs) where one SGP has been ablated by laser-irradiating its ancestor. PGCs are marked with an asterisk. (A) The single (control) SGP is beginning to extend a medial projection posterior to its ispilateral PGC. (B,B′) The two PGCs are in different focal planes. Lobe-like PGC protrusions (typical for this stage) are evident above the cell body. (B) The SGP on the control (un-irradiated) side wraps around its ipsilateral PGC normally, but does not extend to wrap the contralateral PGC (B′). Scale bars are 2.5μm.
PGCs regulate SGP projections and wrapping behaviors but not migration
The interactions we observed between the SGPs and the PGCs suggested that the PGCs might guide SGP morphogenetic behaviors during primordial gonad assembly. In order to test whether the PGCs are required for normal SGP migration behavior, we used mes-1 RNAi, which transforms the PGCs into muscle-like cells that contain P granules within their cytoplasm (Strome et al., 1995). We examined mes-1(RNAi) embryos using 4D fluorescence microscopy and transgenes that mark the SGP and endoderm surfaces, as well as P granules to monitor the effectiveness of mes-1 RNAi. In mes-1(RNAi) embryos, the SGPs were born at the same location as in wild-type embryos (at 48±3% embryo length, n = 6 embryos; Fig. 4D). The SGPs still migrated posteriorly along the lateral edges of the endoderm in mes-1(RNAi) embryos (Fig. 4A–B), and they remained lateral to the endoderm (11/12 SGPs) but ceased migrating at a slightly more anterior position (66±4% embryo length, compared to 70±3% in wild type; n = 14 embryos; Fig. 4D).
In the absence of PGCs, the SGPs extended projections that were abnormal in shape and orientation compared to those in wild-type embryos. While 7/12 SGPs extended projections primarily in a medial direction in mes-1(RNAi) embryos (24/24 SGPs in wild type), 5/12 SGPs extended projections that were abnormally oriented in an anterior, posterior, or ventral direction (Fig.S1). Moreover, the projections in mes-1(RNAi) embryos appeared thicker (arrows in Fig. 4A,B) and more dynamic than those in wild type, and the SGPs extended numerous short projections in various directions (Movie S3). Furthermore, the SGPs in mes-1(RNAi) embryos never wrapped around any cell, and continued to extend projections more than 90 minutes after their birth (0/24 wild-type SGPs were still extending projections at t=90′), even when they came in contact with a transformed (P-granule-containing) muscle cell (Fig. 4C). Welchman et al. (2007) reported that in mes-1(RNAi) larvae, SGPs did not have projections, suggesting that the early dynamic projections eventually collapse. We conclude that PGCs are dispensable for SGP posterior migration but are required for the precise positioning of SGPs at the end of their migration. Further, while PGCs are not needed to initiate formation of SGP projections, PGCs are essential for the morphology and behavior of the SGP projections and for the wrapping behavior of the SGPs. These observations suggest that PGCs provide local signals that organize the SGP projections and promote the transition from the extension of projections (Phase 2) to wrapping behaviors (Phase 3).
Endoderm is required for the morphology and positioning of SGPs at the end of their migration
Since the SGPs migrate in close apposition to the endoderm, we next asked whether endodermal cells regulate SGP morphogenetic behaviors. In end-1 end-3 mutant embryos, the endodermal cell lineage is transformed to adopt epidermal- and muscle-like cell fates (Owraghi et al., 2009). We captured 4D fluorescence movies of end-1 end-3 embryos expressing transgenes that label the SGP surfaces, P granules within PGCs, and transformed endodermal cells (which still express end-1P::mCherry-PHPLC1∂1). We observed that the SGPs were born in approximately the normal location in end-1 end-3 embryos (at 50±4% embryo length, n = 6 embryos; Fig. 4D). However, while the SGPs still migrated posteriorly in the absence of endoderm, they stopped migrating at a more anterior position (65±4% embryo length, compared to 70±3% in wild type; n = 21 embryos; Fig. 4D–G). Since endodermal cells facilitate PGC gastrulation movements, the PGCs remain on the surface of the embryo in many end-1 end-3 mutants (Chihara and Nance, 2012). Notably, the SGPs in end-1 end-3 embryos migrated the same distance posteriorly whether the PGCs were internalized or remained on the embryo’s surface (Fig. 4D).
In the end-1 end-3 embryos in which the PGCs failed to ingress (n = 3 movies), the SGPs extended short dynamic projections in various directions (comparable to the SGPs in mes-1(RNAi) embryos) but did not migrate to the external PGCs nor wrap around any other cell (Fig. 4H). In the end-1 end-3 embryos in which one or both PGCs had ingressed (n = 5 movies), the SGPs always extended projections around the PGCs they contacted, but these projections were often shorter and thicker than in wild type (4/5 movies; arrows in Fig. 4E,F). Furthermore, in end-1 end-3 embryos, one or both SGP cell bodies often shifted more medially than in wild-type embryos (on either the posterior, anterior, or dorsal side of the PGC; 4/5 movies; arrows in Fig. 4F,G), frequently coming into extensive contact with each other (4/5 movies; Fig. 4E–G) and even in temporary contact with the contralateral PGC (3/5 movies; Fig. 4I). However, as long as the PGCs were internal, each SGP eventually wrapped around just its ipsilateral PGC (4/5 movies; in the 5th movie, both SGPs wrapped around the single internalized PGC, as the other PGC remained external, data not shown). We conclude that endoderm is not required for SGPs to migrate posteriorly, extend projections, or wrap around the PGCs. However, endoderm is required for the normal morphology of the SGP projections, for SGPs to reach their regular final position just posterior to the PGCs, and to prevent SGPs from shifting medially and contacting the contralateral SGP and/or PGC. Therefore, the endoderm provides a molecular signal and/or a physical constraint that influences the position and morphology of the SGPs.
To examine the physical constraints provided by surrounding cells during SGP migration, we analyzed transgenic embryos expressing mCherry at all cell membranes and secreted GFP in intercellular spaces (pie-1P::secretedGFP). While the surfaces of most cells in the embryo were contiguous, we found that small spaces opened up between the endoderm and surrounding mesoderm, and the SGPs migrated posteriorly into small pockets of space just lateral to the PGCs (8/8 embryos; Fig. 5A–B). However, we also frequently observed open space just dorsal to the SGPs (arrowhead, Fig. 5A′), yet the SGPs always chose to migrate posteriorly rather than dorsally. Therefore, open space alone is not sufficient to instruct SGP posterior migration, although it could contribute to the final positioning of the SGPs. While the physical landscape provided by the endoderm and PGCs may facilitate SGP positioning, local or global guidance signals likely instruct SGP migration and morphogenetic behavior.
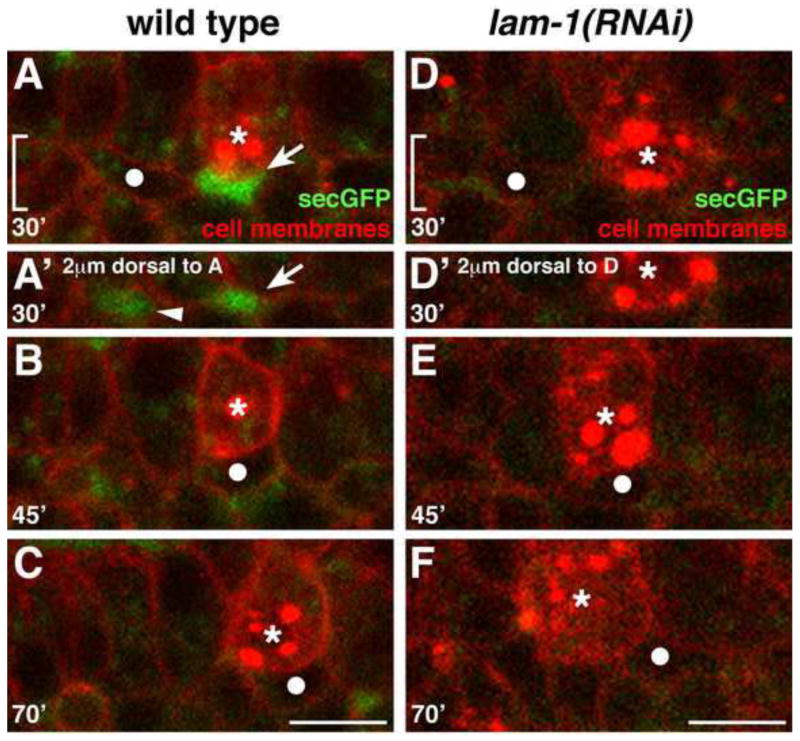
Figure 5. Cell separations are present along the SGP migration route in wild-type but not laminin-depleted embryos.
Stills from a 4D confocal movie of embryo expressing pie-1P::mCherry-PH (cell surfaces), nmy-2P::PGL-1::RFP (PGC-specific P-granules), and pie-1P::secreted GFP (spaces between cells). SGP is indicated with a filled white circle, PGCs with asterisks. Images A–F show a region comparable to that in Fig 1D–F; the midline is at the top of each image. (A–C) Wild-type embryo. (A) Spaces (arrows) can be seen lateral to the endoderm and PGCs, (B–C) and the SGPs migrate into these spaces. (D–F) lam-1(RNAi) embryo. (D–E) Few spaces are evident as the SGPs migrate posteriorly (F) and over-extend past the PGCs. In A′ and D′ the bracketed areas in A and D are shown, at a plane 2μm dorsal to the SGP migration path, to illustrate spaces dorsal to the SGP (arrowhead in A′). Scale bars are 5μm.
Laminin localizes between germ layers as the SGPs migrate and extend projections
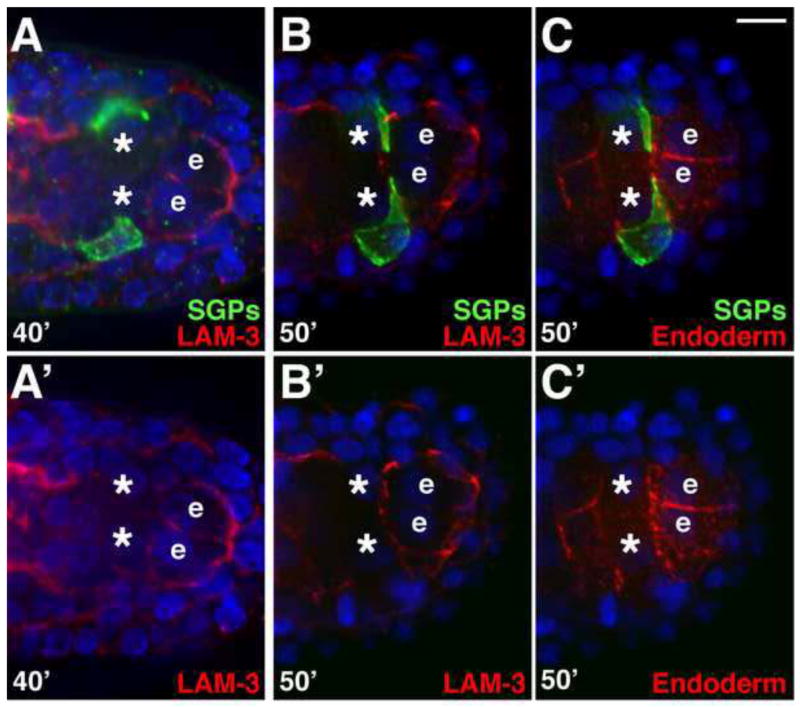
Basement membrane can provide cues that regulate cell migration, the formation of cellular protrusions, and both increased and decreased cell-cell adhesion (Huang et al., 2003; Kao et al., 2006; Mori et al., 2010; reviewed by Yurchenco, 2011). In C. elegans, basement membrane first appears between the germ layers at the middle of embryogenesis, at the approximate stage when the gonadal primordium is forming (Huang et al., 2003; Kao et al., 2006). Because we have shown that the SGPs migrate posteriorly between the endoderm and mesoderm cell layers, we examined the localization of basement membrane relative to the migratory path of the SGPs. Laminin is the first component of the basement membrane to be detected extracellularly (Graham et al., 1997; Huang et al., 2003; Kao et al., 2006), and in other systems, has been shown to organize other basement membrane components (De Arcangelis et al., 1996; Smyth et al., 1999; reviewed by Hohenester and Yurchenco, 2012). Laminin functions as a heterotrimer of α, β, and γ chains, and in C. elegans there are two laminin α chain genes (lam-3 and epi-1) and only single laminin β (lam-1) and laminin γ (lam-2) chain genes (Zhu et al., 2000; Huang et al., 2003; Kao et al., 2006). We examined laminin localization in embryos where the SGP and endoderm surfaces were labeled. At this stage, LAM-3/Laminin αA immunostaining was evident between the germ layers, including on most surfaces of the endoderm (Fig. 6A–C). In particular, LAM-3 localized around the surfaces of the endoderm row 8 cells (labeled “e” in Fig. 6), just before and as the SGPs extended projections between the anterior surfaces of these endoderm cells and the posterior surfaces of the PGCs (Fig. 6A–C). A functional LAM-1-GFP fusion protein showed a similar pattern of localization, including around the gonadal primordium at later stages (asterisk in Fig.S2A). Therefore, laminin basement membrane is present on endodermal surfaces where the SGPs migrate and extend projections around the PGCs.
Figure 6. LAM-3 localizes adjacent to the site of gonadal primordium formation.
Embryos expressing hnd-1P::GFP-PHPLC1∂1 (SGPs) and end-1P::mCherry-PHPLC1∂1 (endoderm), triple-stained for GFP (green), LAM-3 (red in panels A–B′), and mCherry (red in C,C′), as indicated. Nuclei are blue. Images in A′–C′ are single-color panels of corresponding embryos in A–C. B and C are the same embryo. The endodermal row 8 cells are marked (‘e’) and asterisks mark PGCs. (A–A′) Laminin localizes between germ layers and around the endodermal row 8 cells before the SGPs reach the end of their migration, (B–C′) and as the SGPs are extending projections. Brighter LAM-3 on the left side in A–A′ surrounds the pharynx. Scale bar is 5μm.
Laminin is required to prevent SGP over-extension
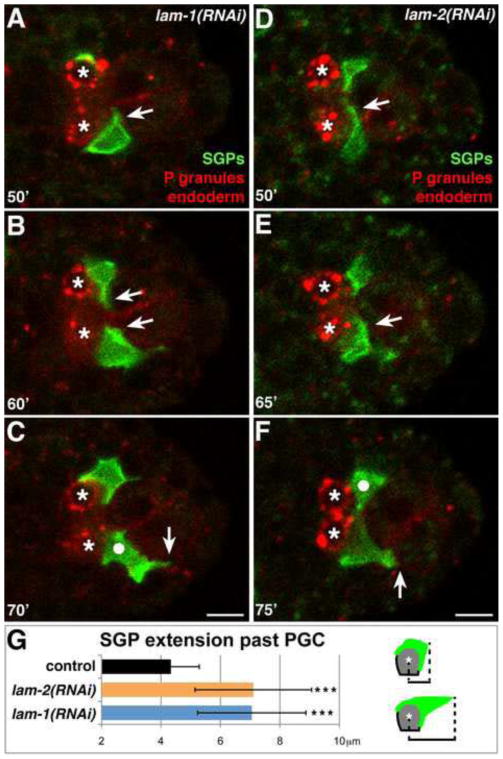
To determine if laminin is required for SGP morphogenesis, we used RNAi to target lam-1/laminin β or lam-2/laminin γ, since LAM-1 and LAM-2 are unique and essential components of the laminin heterotrimer (Huang et al., 2003; Kao et al., 2006). lam-1 RNAi prevented detectable expression of the LAM-1-GFP reporter (Fig.S2B), while lam-2 RNAi effectively blocked secretion of LAM-1-GFP from cells (Fig.S2C), indicating that knockdown of both genes was effective. lam-1(RNAi) embryos and lam-2(RNAi) embryos arrested during late embryogenesis with similar misshapen phenotypes, as observed previously (Kao et al., 2006), but lam-1(RNAi) and lam-2(RNAi) embryos appeared superficially normal at the stage when the primordial gonad formed. We captured 4D movies of lam-1(RNAi) or lam-2(RNAi) embryos expressing SGP and endodermal surface markers as well as the PGC-specific P granule marker (Fig. 7A–F). In lam-1(RNAi) embryos, the SGPs still migrated posteriorly and extended projections around the posterior sides of the PGCs (arrows in Fig. 7A–B; Movie S4). However, while wild-type SGPs always remained lateral to the PGCs and endoderm (24/24 SGPs in 12 movies), SGPs in lam-1(RNAi) embryos often shifted to lie more posterior to the PGCs (13/16 SGPs in 8 movies; circle in Fig. 7C). Most notably, rather than stopping when they reached the PGCs, the SGPs frequently continued extending more posteriorly in lam-1(RNAi) embryos, just lateral to the endodermal row 8 cells (13/16 SGPs in 8 movies; Fig. 7B, arrow in 7C). We observed similar phenotypes in lam-2(RNAi) embryos (Fig. 7D–F). We quantified SGP over-migration by measuring the distance from the center of the PGC nucleus to the posterior-most tip of the SGP at t=70′. While SGPs extended an average of 4±1 μm beyond the PGC nucleus in wild-type embryos (n = 27 embryos), SGPs extended significantly farther posterior in lam-1(RNAi) (n = 20 embryos) and lam-2(RNAi) (n = 43 embryos) embryos, averaging 7±2 μm beyond the PGCs in both genotypes (Fig. 7G). Despite this over-extension, the SGPs always remained in contact with the PGCs, and eventually retracted anteriorly and finished wrapping around the PGCs (16/16 SGPs in 8 movies).
Figure 7. Laminin is required to prevent SGP over-extension past the PGCs.
Stills from a 4D confocal movie of embryo expressing hnd-1P::GFP-PHPLC1∂1 (SGPs), end-1P::mCherry-PHPLC1∂1 (endoderm), and nmy-2P::PGL-1::RFP (PGC-specific P-granules). PGCs are indicated with asterisks. See Fig. 1G–J for comparison to wild type. In lam-1(RNAi) (A–B) and lam-2(RNAi) (D–E) embryos, SGPs extend projections around the PGCs (arrows), (C, F) but SGPs shift to lie posterior to the PGCs (white circles), and over-extend beyond the PGCs (vertical arrows indicate farthest posterior extensions). (G) Quantification of SGP over-extension beyond PGCs in live embryos of the indicated genotype at t=70′. To the right, a schematic of how measurements were taken in representative wild-type (top) and lam-1(RNAi) or lam-2(RNAi) embryos (bottom) is shown. Error bars are standard deviation. (*** = p<0.001, two-tailed Student’s t-test). n = 27, 20, and 43 embryos for control, lam-2(RNAi) and lam-1(RNAi) respectively. Scale bars are 5μm.
Because laminin depletion could affect SGP migration indirectly by altering tissue organization in the embryo, we examined the position of the PGCs and the endoderm in lam-1(RNAi) and in lam-2(RNAi) embryos. We noted small changes in the positioning of the PGCs (Fig.S3A) and the orientation of the endodermal row 8 cells (Fig.S3B,C) in some embryos. However, these phenotypes correlated with SGP over-extension weakly (Fig.S3D) or not at all (Fig.S3E), indicating that laminin likely affects these processes independently. An additional possibility is that laminin depletion creates extra open spaces lateral to the posterior endoderm that the SGPs over-extend into. To test this hypothesis, we examined lam-1(RNAi) embryos expressing markers that label all cell surfaces with mCherry and intercellular spaces with secreted GFP. We found that lam-1(RNAi) embryos did not have any extra pockets of space lateral to the endodermal row 8 cells (compare Fig. 5C to F; n = 8 embryos), and indeed there was little open space around the endoderm at all, including lateral to the PGCs (Fig. 5, compare D–E to A–B). Overall, we conclude that laminin is required to prevent the over-extension of the SGPs, and it is unlikely to do so through alterations in embryo architecture.
DISCUSSION
The C. elegans gonadal primordium provides an excellent model for investigating how stem cell and niche cell precursors first assemble during development. Previous observations of live embryos and larvae revealed the basic organization of the gonadal primordium. For example, in the course of describing the complete embryonic lineage using DIC microscopy, Sulston and colleagues noted that SGPs migrate posteriorly to join the PGCs during mid-embryogenesis (Sulston et al., 1983). Subsequent studies using transgenic strains that mark the cytoplasm of the SGPs with GFP revealed that SGPs appear to cradle the PGCs within the gonadal primordium (Mathies et al., 2003). However, these techniques did not allow observation of the dynamic cellular events and interactions that underlie gonadal primordium assembly. We visualized these steps in detail for the first time by constructing transgenic strains that label the plasma membranes of the SGPs and interacting cells, and by recording time-lapse movies as the gonadal primordium assembles.
Our observations allow us to separate SGP morphogenetic behaviors into three distinct phases (Fig. 1A). In Phase 1, the SGPs migrate posteriorly, along the lateral sides of the endoderm, towards the two PGCs. In Phase 2, each SGP extends a single, long projection around the ipsilateral PGC. In Phase 3, the SGP projections expand over the exposed PGC surfaces, eventually wrapping around the PGCs to form the gonadal primordium. We have determined that neither the endoderm nor the PGCs are required for SGP posterior migration, though both tissues play a role in the final positioning of the SGPs. Further, while neither endoderm nor PGCs are required for the initial extension of SGP projections, both are required for the normal morphology of the projections, and the PGCs are essential for SGP wrapping behavior. Finally, we have shown that laminin localizes between germ layers as the gonadal primordium forms and is required to prevent the SGPs from over-extending past the PGCs at the end of their migration. This work provides a foundation for understanding how the gonadal niche-stem cell interaction is established in C. elegans, and provides a basis for comparison to niche-stem cell assembly events in other species.
SGP posterior migration and final positioning
Because SGPs and PGCs are known to be born in different regions of the embryo in many species, cell migrations are a conserved aspect of gonadogenesis. In Drosophila, the PGCs are first carried into the interior of the embryo by the invaginating endoderm (DeGennaro et al., 2011). PGCs subsequently exit the endoderm and are guided to the somatic gonad precursors through a series of steps, which include attractive cues arising from the SGPs and repulsive cues originating from tissues surrounding the PGC migration path (Starz-Gaiano et al., 2001; Van Doren et al., 1998; Zhang et al., 1997; reviewed by Richardson and Lehmann, 2010). PGC migration in several vertebrate systems also occurs in multiple steps, and migration in zebrafish and mouse is regulated by attractive cues present along the migratory paths of the PGCs and in the location where the gonad will form (reviewed by DeFalco and Capel, 2009; Raz and Reichman-Fried, 2006; Richardson and Lehmann, 2010). Considerably less is known about the morphogenetic movements of the SGPs, but recent work in Drosophila suggests that SGP migration may also be regulated by attractive and repulsive cues from various tissues (Weyers et al., 2011). While there are some differences in the migratory paths and cues involved, the PGCs and SGPs in these model systems must integrate signals from multiple sources as they make their way from their birthplaces to the site of gonad formation.
In C. elegans, the PGCs are internalized passively via association with the endoderm, much like the first phase of Drosophila PGC ingression (Chihara and Nance, 2012). Given the active migration of the SGPs to the PGCs, a reasonable hypothesis would be that the PGCs provide guidance cues that attract the SGPs. However, by genetically transforming the PGCs using mes-1 RNAi, and by observing end-1 end-3 mutants where the PGCs are left on the surface of the embryo, we have shown that the PGCs are not required for the posterior migration of the SGPs. Although the SGPs migrate along the lateral edges of the endoderm, genetic elimination of the endoderm using end-1 end-3 mutants revealed that this tissue also does not play an important role in SGP posterior migration. We therefore speculate that SGP migration cues arise either from another tissue, such as muscle or hypodermis, or from a combination of tissues. Future screens to identify the genes important for SGP morphogenesis will help clarify the source and identity of guidance cues.
Although the endoderm and PGCs are dispensable for SGP posterior migration, both cell types appear to be important for final SGP positioning: when endoderm or PGCs were transformed, the SGPs did not migrate as far posteriorly as in wild type. It is possible that the slight anterior shift in position of SGPs is caused by interference from the transformed cells present in end-1 end-3 or mes-1 mutant embryos, although we noted that the position of transformed cells in both mutants was variable while SGP final position was reproducible. Furthermore, in the absence of endoderm, we noted that SGPs frequently shifted more medially than normal, whereas when PGCs were absent, the SGPs typically remained lateral. These findings suggest that the PGCs induce the SGPs to shift medially, and the endoderm provides a physical block or molecular cues that keep the SGPs lateral. In keeping with this hypothesis, we never observed intercellular spaces between the endoderm cells, suggesting that they adhere tightly to one another and create a physical barrier to medial movement by the SGPs. We suggest that these mechanisms work together to keep the SGPs separated, helping to ensure that each of the PGCs is wrapped by a single SGP.
SGP extension of projections
Although the SGPs extend short protrusions during their posterior migration, upon reaching the PGCs their behavior changes abruptly and each cell extends a long projection over the surface of the adjacent PGC. Long projections are also seen in vivo in some other types of migrating cells, such as Drosophila border cells (Fulga and Rorth, 2002; Prasad and Montell, 2007) and mouse PGCs (Anderson et al., 2000; Blaser et al., 2005; reviewed by Aman and Piotrowski, 2010; Richardson and Lehmann, 2010). In particular, Drosophila SGPs extend long projections around individual PGCs shortly after the cells come into contact, suggesting that this behavior is a conserved aspect of gonadal primordium formation (Jenkins et al., 2003). We similarly observed that SGPs extend long projections after they first come in contact with the PGCs, but we found that these long projections still formed in mes-1(RNAi) embryos. While we cannot exclude the possibility that the transformed PGCs in mes-1(RNAi) embryos retain some characteristics of wild-type PGCs, these data suggest that SGP projections are not induced by the PGCs.
Since the SGP projections extend between the PGCs and endodermal cells, we next hypothesized that endoderm may be required for SGP projections. However, SGPs still extend projections in embryos lacking endoderm (Figure 4E–G). Although SGPs extend their initial projections around the PGCs and towards the midline with mirror-symmetry, we showed that the two SGPs extend projections independently of each other. It is possible that the SGP projections are controlled cell-autonomously. This appears to be the case in zebrafish PGCs, as older PGCs transplanted into younger embryos extend protrusions and migrate to the gonad before endogenous PGCs do (Blaser et al., 2005; reviewed by Aman and Piotrowski, 2010).
Our results also suggest that some extrinsic signals control the directionality and morphology of SGP projections. For example, we showed that in embryos with transformed PGCs, the SGPs frequently extended projections in more than one direction, and only 58% of SGP projections were primarily directed medially, suggesting that PGCs regulate the directionality of SGP projections. In wild-type embryos, it is notable that the SGPs extend a long projection only around the posterior side of the PGCs, even though they first come in contact with the anterior side of the PGCs. However, in embryos with transformed endoderm, the SGPs frequently extended projections around whichever PGC surface they first contacted, suggesting that endoderm may help direct the SGP projections to the posterior side of the PGCs. Additionally, SGP projections appeared shorter and thicker when endoderm or PGCs were absent. Thus, while endoderm and PGCs are not needed for the initiation of SGP projections, they are both required for normal morphology and organization of the projections.
SGP wrapping of the PGCs
A close physical relationship between stem cells and their niches is a conserved and perhaps essential feature of stem cell regulation. For example, in the mammalian testes, the Sertoli cells are tightly associated with the spermatogonial stem cells (SSC), (reviewed by Oatley and Brinster, 2012). Additionally, the SGPs in Drosophila individually ensheathe the PGCs (Van Doren et al., 2003; Jenkins et al., 2003). While the C. elegans gonadal primordium consists of only four cells, C. elegans and Drosophila SGPs carry out a remarkably similar ensheathment of the PGCs. We showed that SGP wrapping is unaffected by the absence of endoderm or the contralateral SGP, but in embryos lacking PGCs, the SGPs never transitioned from the extension of projections (Phase 2) into wrapping behaviors (Phase 3). In Drosophila, SGP ensheathment of PGCs requires the adhesion protein E-cadherin, which is expressed on the surface of both cell types (Jenkins et al., 2003; Li et al., 2003; Van Doren et al., 2003; Weyers et al., 2011). Thus, C. elegans PGCs might also express adhesion proteins on their surfaces that instruct SGP wrapping behavior. Significant depletion of HMR-1/E-cadherin, however, does not prevent C. elegans SGPs from wrapping around PGCs (data not shown), suggesting that alternative adhesion proteins may mediate this interaction in C. elegans.
Laminin and SGP over-extension
The basement membrane component laminin plays a role in many different morphogenetic processes, including directing cell migrations, inducing cell projections, regulating adhesion, and polarizing tissues (Huang et al., 2003; Kao et al., 2006; Mori et al., 2010; Rasmussen et al., 2012; reviewed by Yurchenco, 2011). In Drosophila, laminin mutations result in the misplacement of the gonad, or of individual PGCs or SGP clusters (Jaglarz and Howard, 1995; Weyers et al., 2011). In mouse, laminin is expressed broadly in the embryo, including along the path of PGC migration, and PGCs end their migration at a region of heightened laminin (Garcia-Castro et al., 1997). We observed that laminin localized between germ layers at the time the SGPs migrate posteriorly (see also Huang et al., 2003; Kao et al, 2006). In particular, we noticed that laminin was always present around the surfaces of the endodermal row 8 cells, which the SGPs contact as they extend their long projections and cease migration. Although laminin was not required for SGP posterior migration or extension of projections, we did find that laminin depletion caused SGPs to over-extend significantly beyond the PGCs.
How does laminin depletion affect SGP protrusions? Since laminin mutations cause defects in cell adhesion in other tissues in C. elegans (Huang et al., 2003), one possibility is that laminin depletion disrupts cell-cell adhesion around the posterior endoderm and thus allows the SGPs to extend farther posteriorly, lateral to these cells. However, when we examined intercellular spaces in lam-1(RNAi) embryos, we observed even fewer open spaces, suggesting that SGP over-extension is not the result of reduced cell-cell adhesion. Basement membrane can also play a role in cell-cell signaling, either directly via laminin receptors, or indirectly by trapping and thus concentrating secreted signals (Wang et al., 2008; reviewed by Yurchenco, 2011). Laminin depletion could therefore alter the secretion or distribution of a protein that guides SGP projections. In wild-type embryos, the SGPs do not stop migrating when they first come in contact with the PGCs. Rather, they stop only after extending a long projection around the posterior surface of the PGCs, where we observed laminin localization between the PGCs and endoderm. This region of laminin expression may directly signal the SGPs to stop extending posteriorly, or it may trap a “stop” signal released by neighboring tissues. Since the mechanisms through which laminin functions in Drosophila and mouse gonad formation are also not yet known, it will be interesting to learn if laminin functions similarly in each species.
Conclusions
Stem cells are critical for the development and homeostasis of many tissues, and the regulation provided by the niche is critical for stem cell survival and proliferation (reviewed by Hubbard, 2007; Byrd and Kimble, 2009; Lehmann, 2012; Losick et al., 2011). Interactions and communication between niche cells and stem cells has been studied most extensively in the gonad of several model systems, including Drosophila, C. elegans, and mouse (reviewed by Hubbard, 2007; Lehmann, 2012; Losick et al., 2011; Morrison and Spradling, 2008). Gonadal development is a complex but critical process, as SGPs and PGCs respond to a wide variety of signals as they migrate through the embryo and finally coalesce to initiate the niche-stem cell relationship. Research in Drosophila, mouse, and zebrafish has established some conserved principles of gonadal development, including cell migrations and SGP extension of projections and wrapping of the PGCs. Although it may be the exception for SGPs to migrate more extensively than PGCs, the migration of the C. elegans SGPs shares many conserved elements with PGC migration in other species, in that it is a multi-step process that requires signals from several tissues to guide the cells to their final position. While the coalescence of the somatic and germ cells is not as well understood in vertebrates, both Drosophila and C. elegans SGPs extend long projections around the PGCs, eventually ensheathing them to form the gonadal primordium. As there are few systems in which these events are well understood in real time, our findings contribute to an overall understanding of the morphogenetic events of gonadal development.
The mechanisms that control gonadal development in C. elegans may have implications in other systems as well. Hematopoietic stem cells are also born away from their niche, and their migration to the niche appears to be regulated by some of the same genes that regulate gonadal development (reviewed by Boisset and Robin, 2012; Mazo et al., 2011; Morrison and Spradling, 2008). And in C. elegans, the SGPs share a similar wrapping morphology with their granddaughter cells, the somatic DTCs, which wrap around the distal end of each gonad arm and provide a niche for the germline stem cells (reviewed by Hubbard, 2007). It is interesting to speculate that the molecular mechanisms controlling these behaviors might also be shared. Regardless, the early establishment of a close interaction between the somatic and germ cells highlights the importance of the relationship between the stem cells and their niche. The study of C. elegans gonadal development will provide important insights into our overall understanding of gonadal development, as well as potential insights into other niche-stem cell interactions that involve cell migrations or close adhesion between niche cells and stem cells.
Supplementary Material
Highlights.
We visualize gonadal promordium assembly using timelapse microscopy
We identify three phases of SGP behavior: migration, projection extension, wrapping
We show that endoderm and PGCs help position SGPs, and PGCs instruct wrapping
We find that laminin keeps SGPs from overextending beyond the PGCs.
Acknowledgments
We thank Julie Ahringer, Morris Maduro, James Priess, Jeff Rasmussen, and William Wadsworth for their generous gifts of strains and reagents, and Jane Hubbard, Jessica Seifert, and members of the Nance and Hubbard labs for helpful discussions and comments on the paper. We thank Daisuke Chihara for assistance in carrying out laser ablations. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD10440). Funding was provided by an ACS Postdoctoral Fellowship (M.R.R.) and NIH grant R21HD058953 (J.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Anderson DC, Gill JS, Cinalli RM, Nance J. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 2008;320:1771–1774. doi: 10.1126/science.1156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R, Copeland TK, Scholer H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech Dev. 2000;91:61–68. doi: 10.1016/s0925-4773(99)00271-3. [DOI] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, 3rd, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H, Eisenbeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, Raz E. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J Cell Sci. 2005;118:4027–4038. doi: 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- Boisset JC, Robin C. On the origin of hematopoietic stem cells: progress and controversy. Stem Cell Res. 2012;8:1–13. doi: 10.1016/j.scr.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Byrd DT, Kimble J. Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin Cell Dev Biol. 2009;20:1107–1113. doi: 10.1016/j.semcdb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara D, Nance J. An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation. Development. 2012;139:2547–2556. doi: 10.1242/dev.079863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arcangelis A, Neuville P, Boukamel R, Lefebvre O, Kedinger M, Simon-Assmann P. Inhibition of laminin alpha 1-chain expression leads to alteration of basement membrane assembly and cell differentiation. J Cell Biol. 1996;133:417–430. doi: 10.1083/jcb.133.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T, Capel B. Gonad morphogenesis in vertebrates: divergent means to a convergent end. Annu Rev Cell Dev Biol. 2009;25:457–482. doi: 10.1146/annurev.cellbio.042308.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro M, Hurd TR, Siekhaus DE, Biteau B, Jasper H, Lehmann R. Peroxiredoxin stabilization of DE-cadherin promotes primordial germ cell adhesion. Dev Cell. 2011;20:233–243. doi: 10.1016/j.devcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Rorth P. Invasive cell migration is initiated by guided growth of long cellular extensions. Nat Cell Biol. 2002;4:715–719. doi: 10.1038/ncb848. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Anderson R, Heasman J, Wylie C. Interactions between germ cells and extracellular matrix glycoproteins during migration and gonad assembly in the mouse embryo. J Cell Biol. 1997;138:471–480. doi: 10.1083/jcb.138.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, Kramer JM. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J Cell Biol. 1997;137:1171–1183. doi: 10.1083/jcb.137.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenester E, Yurchenco PD. Laminins in basement membrane assembly. Cell Adh Migr. 2012:7. doi: 10.4161/cam.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Hall DH, Hedgecock EM, Kao G, Karantza V, Vogel BE, Hutter H, Chisholm AD, Yurchenco PD, Wadsworth WG. Laminin alpha subunits and their role in C. elegans development. Development. 2003;130:3343–3358. doi: 10.1242/dev.00481. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ. Caenorhabditis elegans germ line: a model for stem cell biology. Dev Dyn. 2007;236:3343–3357. doi: 10.1002/dvdy.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<2::AID-DVDY2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Jaglarz MK, Howard KR. The active migration of Drosophila primordial germ cells. Development. 1995;121:3495–3503. doi: 10.1242/dev.121.11.3495. [DOI] [PubMed] [Google Scholar]
- Jenkins AB, McCaffery JM, Van Doren M. Drosophila E-cadherin is essential for proper germ cell-soma interaction during gonad morphogenesis. Development. 2003;130:4417–4426. doi: 10.1242/dev.00639. [DOI] [PubMed] [Google Scholar]
- Kao G, Huang CC, Hedgecock EM, Hall DH, Wadsworth WG. The role of the laminin beta subunit in laminin heterotrimer assembly and basement membrane function and development in C. elegans. Dev Biol. 2006;290:211–219. doi: 10.1016/j.ydbio.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Lehmann R. Germline stem cells: origin and destiny. Cell Stem Cell. 2012;10:729–739. doi: 10.1016/j.stem.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- Losick VP, Morris LX, Fox DT, Spradling A. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell. 2011;21:159–171. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies LD, Henderson ST, Kimble J. The C. elegans Hand gene controls embryogenesis and early gonadogenesis. Development. 2003;130:2881–2892. doi: 10.1242/dev.00483. [DOI] [PubMed] [Google Scholar]
- Mazo IB, Massberg S, von Andrian UH. Hematopoietic stem and progenitor cell trafficking. Trends Immunol. 2011;32:493–503. doi: 10.1016/j.it.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol. 2008;18:1476–1482. doi: 10.1016/j.cub.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Ono K, Kariya Y, Ogawa T, Higashi S, Miyazaki K. Laminin-3B11, a novel vascular-type laminin capable of inducing prominent lamellipodial protrusions in microvascular endothelial cells. J Biol Chem. 2010;285:35068–35078. doi: 10.1074/jbc.M110.146126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol Rev. 2012;92:577–595. doi: 10.1152/physrev.00025.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev Biol. 2010;340:209–221. doi: 10.1016/j.ydbio.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Rasmussen JP, Reddy SS, Priess JR. Laminin is required to orient epithelial polarity in the C. elegans pharynx. Development. 2012;139:2050–2060. doi: 10.1242/dev.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E, Reichman-Fried M. Attraction rules: germ cell migration in zebrafish. Curr Opin Genet Dev. 2006;16:355–359. doi: 10.1016/j.gde.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottach A, Kremmer E, Nowak D, Leonhardt H, Cardoso MC. Generation and characterization of a rat monoclonal antibody specific for multiple red fluorescent proteins. Hybridoma (Larchmt) 2008;27:337–343. doi: 10.1089/hyb.2008.0031. [DOI] [PubMed] [Google Scholar]
- Smyth N, Vatansever HS, Murray P, Meyer M, Frie C, Paulsson M, Edgar D. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J Cell Biol. 1999;144:151–160. doi: 10.1083/jcb.144.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starz-Gaiano M, Cho NK, Forbes A, Lehmann R. Spatially restricted activity of a Drosophila lipid phosphatase guides migrating germ cells. Development. 2001;128:983–991. doi: 10.1242/dev.128.6.983. [DOI] [PubMed] [Google Scholar]
- Strome S, Martin P, Schierenberg E, Paulsen J. Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development. 1995;121:2961–2972. doi: 10.1242/dev.121.9.2961. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Broihier HT, Moore LA, Lehmann R. HMG-CoA reductase guides migrating primordial germ cells. Nature. 1998;396:466–469. doi: 10.1038/24871. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Mathews WR, Samuels M, Moore LA, Broihier HT, Lehmann R. fear of intimacy encodes a novel transmembrane protein required for gonad morphogenesis in Drosophila. Development. 2003;130:2355–2364. doi: 10.1242/dev.00454. [DOI] [PubMed] [Google Scholar]
- Wang X, Harris RE, Bayston LJ, Ashe HL. Type IV collagens regulate BMP signalling in Drosophila. Nature. 2008;455:72–77. doi: 10.1038/nature07214. [DOI] [PubMed] [Google Scholar]
- Welchman DP, Mathies LD, Ahringer J. Similar requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in diverse cell types. Dev Biol. 2007;305:347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers JJ, Milutinovich AB, Takeda Y, Jemc JC, Van Doren M. A genetic screen for mutations affecting gonad formation in Drosophila reveals a role for the slit/robo pathway. Dev Biol. 2011;353:217–228. doi: 10.1016/j.ydbio.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser E, Frokjaer-Jensen C, Jorgensen E, Ahringer J. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS One. 2011;6:e20082. doi: 10.1371/journal.pone.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhang J, Purcell KJ, Cheng Y, Howard K. The Drosophila protein Wunen repels migrating germ cells. Nature. 1997;385:64–67. doi: 10.1038/385064a0. [DOI] [PubMed] [Google Scholar]
- Zhu X, Joh K, Hedgecock EM, Hori K. Identification of epi-1 locus as a laminin alpha chain gene in the nematode Caenorhabditis elegans and characterization of epi-1 mutant alleles. DNA Seq. 1999;10:207–217. doi: 10.3109/10425179909033950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.