Figure 7.
DDK at Kinetochores Independently Regulates Replication Timing and Cohesion at Pericentromeres
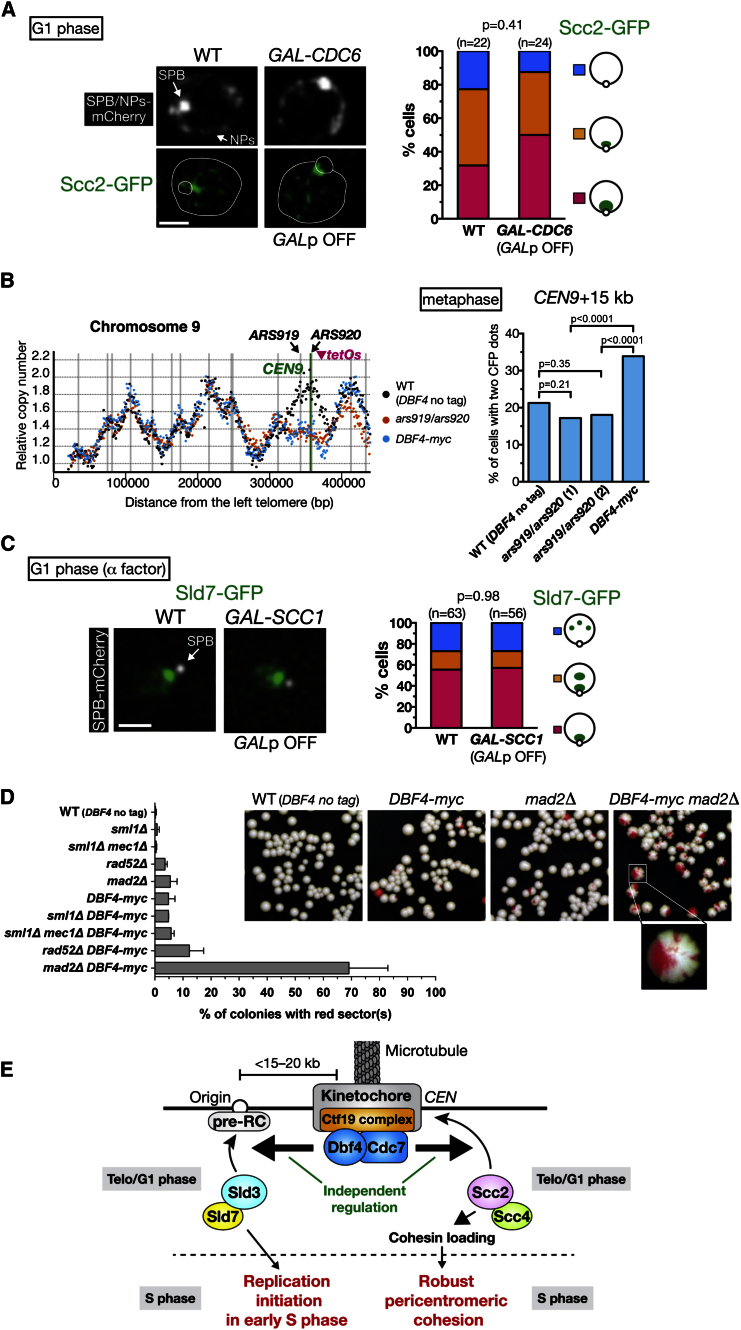
(A) Scc2 accumulation at centromeric regions does not require the pre-RC in G1 phase. CDC6+ (WT, T10558) and GAL-CDC6 (T10559) cells with SCC2-GFP SPC42-mCherry NIC96-mCherry were treated as in Figure 2Di. The scale bar represents 1 μm.
(B) Inactivation of CEN9-proximal origins does not weaken cohesion at the CEN9 pericentromere. WT (DBF4 no tag, T10461, n = 282), ars919/ars920 (clone 1, T10552, n = 314; clone 2, T10770, n = 316), and DBF4-myc (T10528, n = 295) cells with tetOs at +15 kb from CEN9 were treated as in Figure 5A and analyzed at 160 min after release from α factor. Left: replication timing of chromosome 9.
(C) Cohesin is not required for Sld7 accumulation at centromeric regions in G1 phase. SCC1+ (WT, T9110) and GAL-SCC1 (T9107) cells with SLD7-GFP SPC42-mCherry were analyzed as in Figure 2Ci.
(D) DBF4-myc cells show an increase in chromosome loss (p = 0.0006), which is further enhanced by deletion of MAD2 (p < 0.0001). Indicated strains with the CFIII chromosome fragment were used for a chromosome loss assay. Loss of CFIII generated red sector in colonies. Error bars represent SD.
(E) Summary of two functions of DDK recruited to kinetochores. See details in the text.
See also Figure S7.