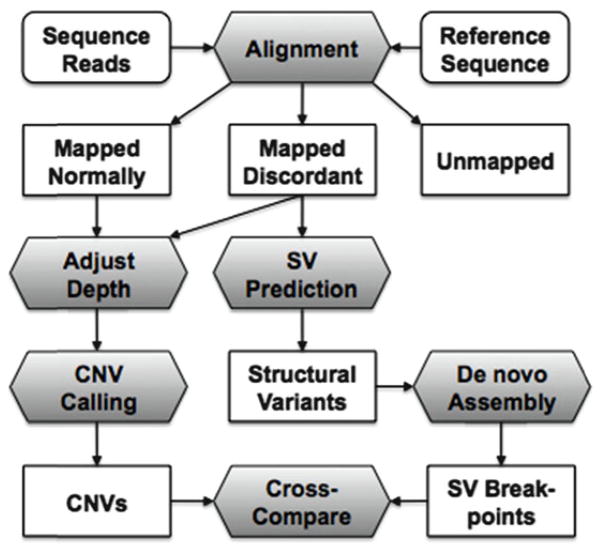
Fig. 1.
Detection of copy number variation (CNV) and structural variation (SV) by massively parallel sequencing. When aligned to a reference sequence, read pairs are classified as mapped normally, mapped discordantly, or unmapped. Both classes of mapped reads are used for inference of read depth and then CNV calling. Discordant pairs are utilized for prediction of SVs, which are further resolved by de novo assembly using both discordant and unmapped reads.