Fig. 2.
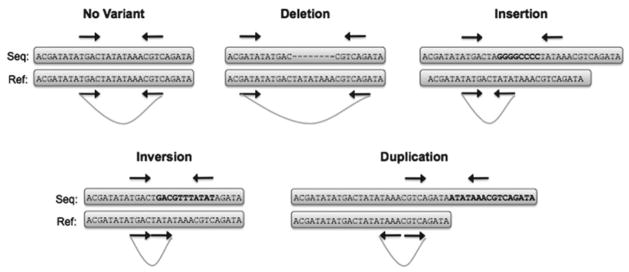
Detecting structural variants by paired-end mapping. The distance between and relative orientation of associated read pairs suggests specific classes of SVs. Deletions produce reads that map more distant from one another than expected while insertions have the opposite effect. Read pairs spanning breakpoints of inversions and duplications have altered distance and orientation while read pairs spanning translocations (not shown) will map to different chromosomes.
