Figure 4.
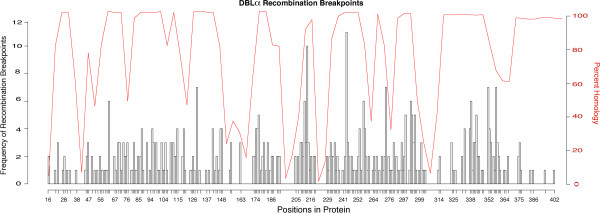
Our analysis shows that recombination is uniform throughout the DBLα domains and does not show a hot- or coldspot structure. This lack of structure is revealed in this histogram showing recombination breakpoint frequency at each amino acid position mapped onto a multiple sequence alignment (white bars). This is overlaid with levels of homology (percent of gaps at each position) at each site (red line). Levels of homology are highly inconsistent over the length of DBLα domains and regions that are highly homologous and align well have no gaps, whereas regions with low homology require many gaps. This figure shows that, correcting for regions where there are large gaps (low homology), there are no large increases or decreases in frequency of recombination breaks typical of hot- or coldspot structure. The multiple sequence alignment is of all DBLα domains from all samples used in this study.
