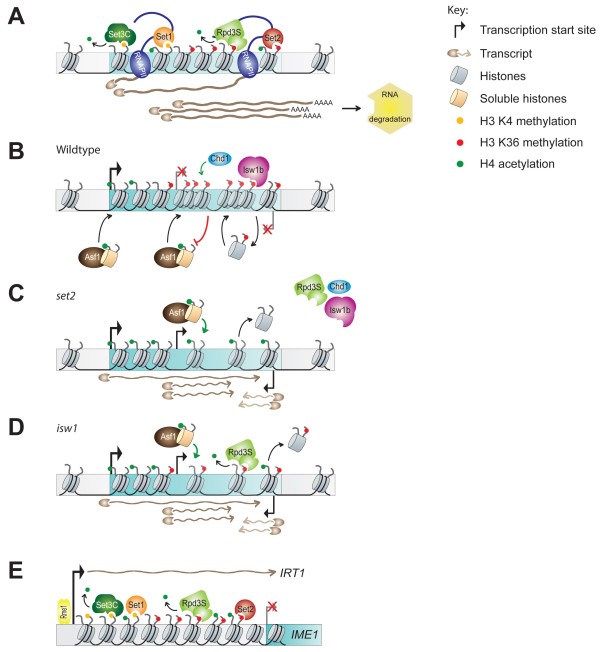
Figure 2.
Organized chromatin structure antagonizes production of ncRNAs. (A) The RNAPII-associated KMTases Set1 and Set2 are required to methylate histone H3 on residues K4 and K36, respectively. H3 K4 dimethylation and H3 K36 trimethylation are essential for maintaining coding regions in a hypoacetylated state: H3 K4me2 directly recruits the histone deacetylase complex Set3C, while H3 K36 methylation is essential for the catalytic activity of the Rpd3S deacetylase complex. (B) H3 K36 methylation directly recruits the Isw1b remodeling complex through a PWWP domain in its Ioc4 subunit. Isw1b together with Chd1 are necessary for the retention of existing, H3 K36 methylated nucleosomes over coding regions. Thereby they prevent deposition of soluble, highly acetylated histones through histone chaperones such as Asf1. (C) Loss of SET2 completely abolishes H3 K36 methylation in yeast. Isw1b is no longer correctly recruited to chromatin, resulting in increased histone exchange with a concomitant rise in histone acetylation over coding sequences. This leads to the exposure of cryptic promoters and the production of ncRNAs. (D) Loss of either ISW1 and/or CHD1 also results in increased histone exchange and histone acetylation even though there is little change in H3 K36me3 levels over the gene body. However, in the absence of the remodeler(s) existing nucleosomes cannot be retained. Instead they are replaced with soluble, highly acetylated histones. Again this process leads to the exposure of cryptic promoters and the production of ncRNAs. (E) In haploid yeast Rme1-dependent transcription of the long ncRNA IRT1 establishes a gradient of H3 K4me2 and H3 K36me2/3 over the promoter of the IME1 gene. These methylation marks subsequently recruit histone deacetylases Set3C and Rpd3S, respectively, that help to establish a repressive chromatin conformation and thus preclude transcription of IME1.