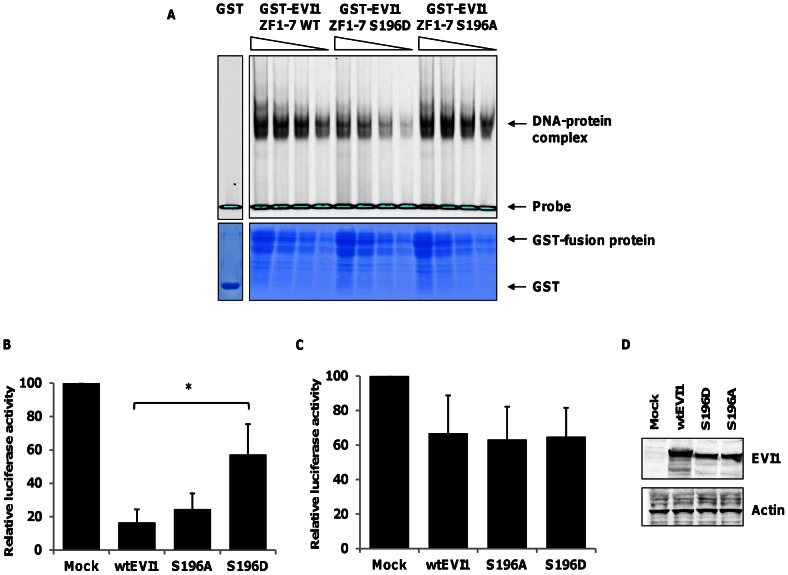
Figure 3. EVI1 S196 phosphorylation disrupts DNA binding and transcriptional repression.
(A) Electro-mobility shift assay (EMSA) with GST alone or GST-Evi1 (ZF1-7) fusion proteins corresponding to WT or mutant Evi1 mimicking non-phosphorylated (S196A) or phosphorylated serine 196 (S196D) (upper panel). The DNA-protein complex and the free, labelled DNA probe are indicated. Lower panel: GST-fusion proteins separated on a 12.5% (w/v) SDS-PAGE gel and stained with colloidal coomassie blue. Specificity of protein/DNA interaction was confirmed by incubation with excess concentration of unlabelled random DNA substrate (data not shown). Graphs show mean (±s.d.,n = 4) luciferase activity relative to an empty vector control from reporter gene assays using either (B) a PLZF-luc reporter or (C) a Fos reporter co-transfected into HEK 293 cells with wild-type Evi1, S196 mutants or empty vector control. Luciferase activities were normalised against Renilla activity and calibrated to the empty vector control, *P<0.01, ns = non-significant (D) Western blotting of wild-type and mutant Evi1 protein levels in the reporter gene assays.