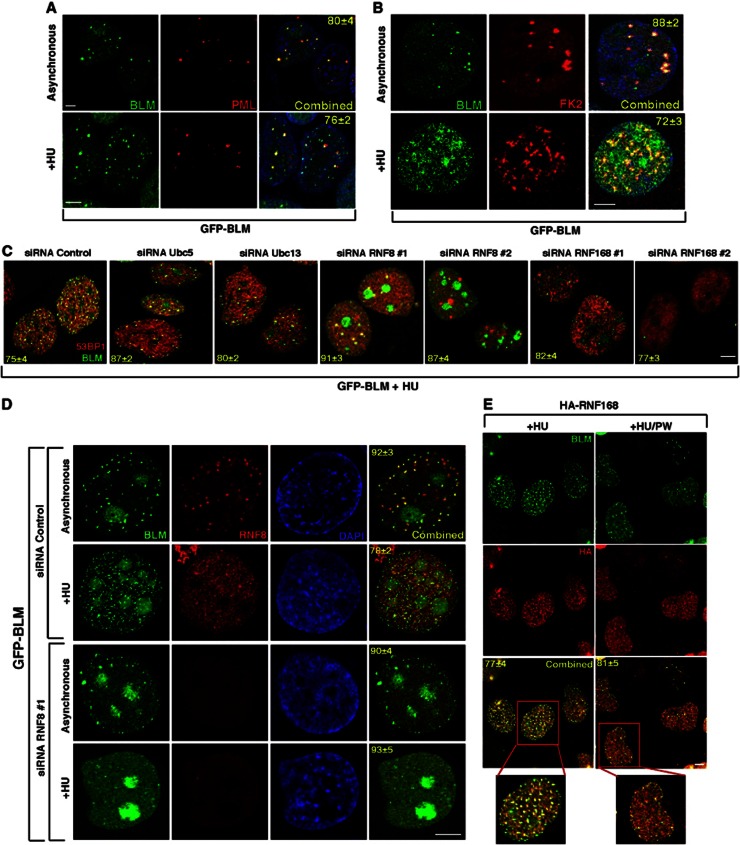
Figure 1.
Recruitment of BLM to the sites of stalled replication depends on RNF8 and RNF168. (A, B) BLM colocalizes with PML-NBs and ubiquitin in the absence of DNA damage. GFP-BLM cells were either grown under asynchronous conditions or treated with HU (+HU). GFP-BLM cells were co-stained with (A) anti-PML antibody and (B) anti-ubiquitin FK2 antibody. Nuclei are stained by DAPI. Scale 5 μM. (C) Lack of E3 ligases leads to lack of BLM recruitment after HU treatment. GFP-BLM cells were transfected with either the control siRNA or siRNAs against Ubc5, Ubc13, RNF8 or RNF168. The cells were treated with HU. GFP-BLM was visualized along with 53BP1. Scale 5 μM. (D) BLM accumulates in the nucleolus in the absence of RNF8. Same as (C) except only siRNA Control or siRNA RNF8 #1 was used. GFP-BLM cells were stained with anti-RNF8 antibodies. Nuclei visualized by DAPI. Scale 5 μM. (E) RNF168 colocalizes with BLM after HU treatment. RIDDLE cells complemented with HA-tagged RNF168 were grown in the presence of HU (+HU) or in the postwash condition (+HU/PW). One of the cells under either conditions is zoomed. Immunofluorescence was carried out with anti-BLM (A300–110A) and anti-HA antibodies. Scale 5 μM.