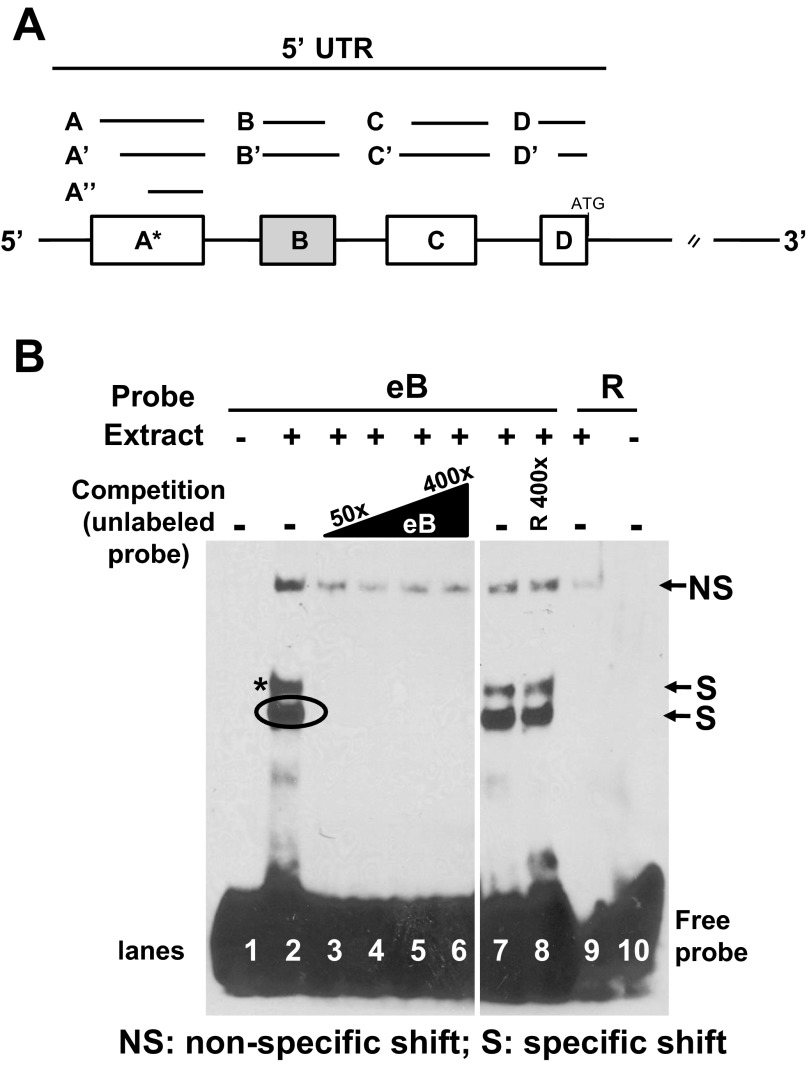
Fig. 1.
5′ Untranslated region (5′UTR) structure of SP-A1 and SP-A2 and exon B (eB) sequence-specific shifts. A: exons A, B, C, D differ in length and are represented by lines and boxes. The sizes of each untranslated exon are as follows: A (44 nt), A′ (39 nt), A″ (34 nt), B (30 nt), B′ (70 nt), C (60 nt), C′ (63 nt), D (26 nt), and D′ (23 nt). Alternative splicing of 5′UTR exons gives rise to several SP-A1 and SP-A2 variants; the most frequent variant for SP-A1 is AD′ and for SP-A2 are ABD and ABD′. B: RNA electromobility shift assays (REMSA) of eB biotinylated probe with cytoplasmic extract of NCI-H441 cells. Lane 1, free probe (negative control); lanes 2 and 7, 200 nM eB riboprobe incubated with 8.5 μg extract (positive controls); lanes 3–6, competition of eB-mediated shifts with 50-, 100-, 200-, and 400-fold molar excess of unlabeled eB RNA, respectively; lane 8, competition of eB shifts with 400-fold molar excess of unlabeled random (R) RNA; lane 9, R biotinylated probe incubated with extract; lane 10, R probe; (+) presence of NCI-H441 cytoplasmic extract; (−) absence of cytoplasmic extract or competitor RNA. The top shift band is a nonspecific shift (NS) band because it appears in the random probe (R) (lane 9) and is not competed. Lanes 1–10 are from the same film but some lanes between lanes 6 and 7 were removed.