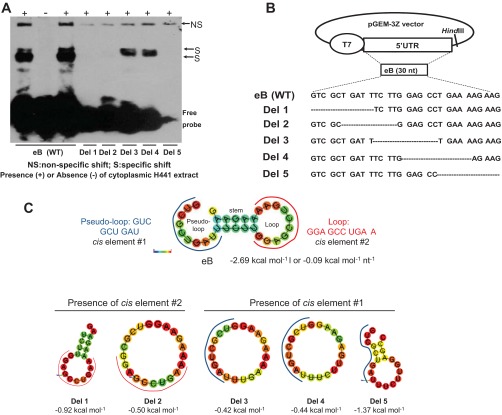
Fig. 2.
Deletion mapping of eB cis-acting elements and predicted secondary structure of WT eB and deletion mutant mRNAs. A: REMSA results of biotinylated wild-type (WT) and deletion (Del) mutant riboprobes with cytoplasmic extract of NCI-H441 cells. Del 3 and Del 4 formed sequence-specific shifts whereas the rest deletion mutants did not. B: the WT eB sequence and the sequences of the deletion mutants. The mutants were generated by site-directed mutagenesis in pGEM-3Z vector by sequentially removing 10 nt. C: the secondary structure of WT eB and deletion mutants, as well as the sequence and location of the two eB cis-elements. The eB secondary structure is described by a pseudo-loop, a stem, and a loop. The secondary structure of Del 5 differs from that of Del 3 and Del 4. In particular, the secondary structures of both Del 3 and Del 4 are single loops whereas Del 5 results in a small loop, a short stem, and a short pseudo-loop. The RNAfold online software was used to obtain the secondary structure of WT eB and deletion mutants. The minimum free energy secondary structure was used, with the best possible combination of paired bases, and the minimum free energy (ΔG) normalized for size, by dividing ΔG by the total number of bases of each experimental construct. The colors indicate the propensity of individual nucleotides to participate in base pairs. The scale ranges from red (highest probability) to blue-violet (lower probability).