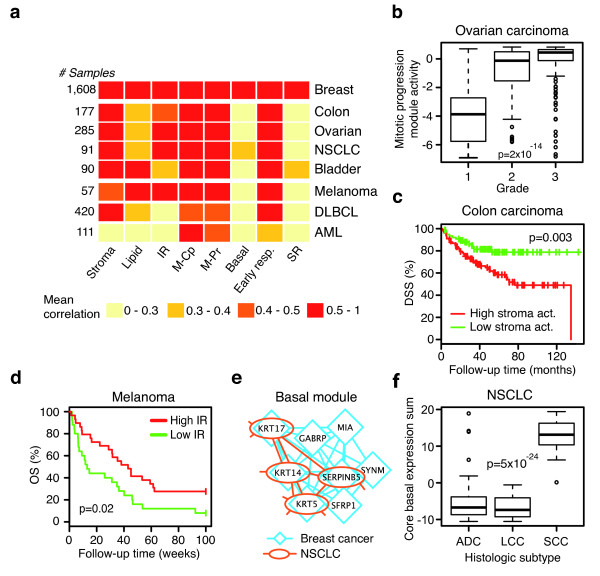
Figure 4.
The breast cancer-derived gene expression modules are preserved across several cancer forms. (a) The breast cancer gene expression modules were assayed for co-expression in data representing seven other cancer forms by calculating the average pair-wise Pearson correlation for genes within each module separately. All observed correlations were significant as compared to a random average pair-wise correlations based on 1,000 permutations (data not shown) M-Pr, mitotic progression; M-Cp, mitotic checkpoint. (b) A high activity score of the mitotic progression module correlated to increasing grade in an ovarian carcinoma dataset (n = 285, P = 2 × 10-14, ANOVA). (c) An above mean expression of genes in the stroma module correlates to decreased disease-specific survival (DSS) in a colon carcinoma dataset. (n = 177, P = 0.003, log-rank test). (d) A high immune response (IR) module activity correlated to favorable overall survival (OS) in a dataset representing 57 stage IV melanomas (P = 0.02, log-rank test). (e) Calculation of pair-wise Pearson correlations in an NSCLC dataset for genes in the breast cancer basal module (blue network) revealed that only a subset of these genes were correlated in NSCLC (red network). A core basal gene expression module (n = 5) was derived from genes with conserved correlations in both breast and lung cancer data (red network). (f) A high expression sum for the core basal module acted as a marker for squamous cell lung carcinoma (SCC) compared to the other NSCLC morphological types adenocarcinoma (ADC) and large cell carcinoma (LCC) (P = 5*10-24, ANOVA).