Abstract
Transcription of the Kluyveromyces lactis beta-galactosidase gene, LAC4, is inducible by galactose and lactose. We examined the effects of deletion mutations within the LAC4 promoter on the expression of beta-galactosidase activity. The results of these experiments indicate that at least two upstream activator sequences (UAS) mediate maximum induction by galactose. These UAS sequence elements are homologous to UAS that regulate induction of the melibiose-galactose regulon of Saccharomyces cerevisiae. We also show that a synthetic copy of one of the K. lactis UAS restores the inducibility of a deleted, noninducible LAC4 promoter. Since the uninduced or basal level of LAC4 expression was increased in several promoter deletion strains and in deletion strains carrying one or two synthetic UAS, we examined the contribution of the LAC9 positive regulatory protein to this effect. The LAC9 protein is thought to bind to UAS and activate transcription of LAC4 (L.V. Wray, M.M. Witte, R.C. Dickson, and M.I. Riley, Mol. Cell. Biol. 7:1111-1121, 1987). Our results demonstrate that LAC9 protein plays a role in setting the uninduced level of gene expression, but other factors also participate. For example, in a lac9 background a LAC4 promoter deletion mutant with two copies of a synthetic 17-base-pair UAS yields a sevenfold higher level of uninduced LAC4 expression than the same strain with one UAS. These and other data indicate that the basal level of gene expression is strongly influenced by the base sequence of the promoter.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
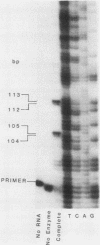
- Breunig K. D., Dahlems U., Das S., Hollenberg C. P. Analysis of a eukaryotic beta-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res. 1984 Mar 12;12(5):2327–2341. doi: 10.1093/nar/12.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- DOUGLAS H. C., HAWTHORNE D. C. ENZYMATIC EXPRESSION AND GENETIC LINKAGE OF GENES CONTROLLING GALACTOSE UTILIZATION IN SACCHAROMYCES. Genetics. 1964 May;49:837–844. doi: 10.1093/genetics/49.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Breunig K. D., Hollenberg C. P. A positive regulatory element is involved in the induction of the beta-galactosidase gene from Kluyveromyces lactis. EMBO J. 1985 Mar;4(3):793–798. doi: 10.1002/j.1460-2075.1985.tb03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Molecular cloning and expression in E. coli of a yeast gene coding for beta-galactosidase. Cell. 1978 Sep;15(1):123–130. doi: 10.1016/0092-8674(78)90088-0. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Sheetz R. M., Lacy L. R. Genetic regulation: yeast mutants constitutive for beta-galactosidase activity have an increased level of beta-galactosidase messenger ribonucleic acid. Mol Cell Biol. 1981 Nov;1(11):1048–1056. doi: 10.1128/mcb.1.11.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas H. C., Hawthorne C. D. Uninducible mutants in the gal i locus of Saccharomyces cerevisiae. J Bacteriol. 1972 Mar;109(3):1139–1143. doi: 10.1128/jb.109.3.1139-1143.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper J. E., Broach J. R., Rowe L. B. Regulation of the galactose pathway in Saccharomyces cerevisiae: induction of uridyl transferase mRNA and dependency on GAL4 gene function. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2878–2882. doi: 10.1073/pnas.75.6.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy L. R., Dickson R. C. Transcriptional regulation of the Kluyveromyces lactis beta-galactosidase gene. Mol Cell Biol. 1981 Jul;1(7):629–634. doi: 10.1128/mcb.1.7.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Riley M. I., Dickson R. C. Genetic and biochemical characterization of the galactose gene cluster in Kluyveromyces lactis. J Bacteriol. 1984 May;158(2):705–712. doi: 10.1128/jb.158.2.705-712.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph H., Koenig-Rauseo I., Hinnen A. One-step gene replacement in yeast by cotransformation. Gene. 1985;36(1-2):87–95. doi: 10.1016/0378-1119(85)90072-1. [DOI] [PubMed] [Google Scholar]
- Sheetz R. M., Dickson R. C. Lac4 is the structural gene for beta-galactosidase in Kluyveromyces lactis. Genetics. 1981 Aug;98(4):729–745. doi: 10.1093/genetics/98.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz R. M., Dickson R. C. Mutations affecting synthesis of beta-galactosidase activity in the yeast Kluyveromyces lactis. Genetics. 1980 Aug;95(4):877–890. doi: 10.1093/genetics/95.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sreekrishna K., Dickson R. C. Construction of strains of Saccharomyces cerevisiae that grow on lactose. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7909–7913. doi: 10.1073/pnas.82.23.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekrishna K., Webster T. D., Dickson R. C. Transformation of Kluyveromyces lactis with the kanamycin (G418) resistance gene of Tn903. Gene. 1984 Apr;28(1):73–81. doi: 10.1016/0378-1119(84)90089-1. [DOI] [PubMed] [Google Scholar]
- Sures I., Levy S., Kedes L. H. Leader sequences of Strongylocentrotus purpuratus histone mRNAs start at a unique heptanucleotide common to all five histone genes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1265–1269. doi: 10.1073/pnas.77.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]