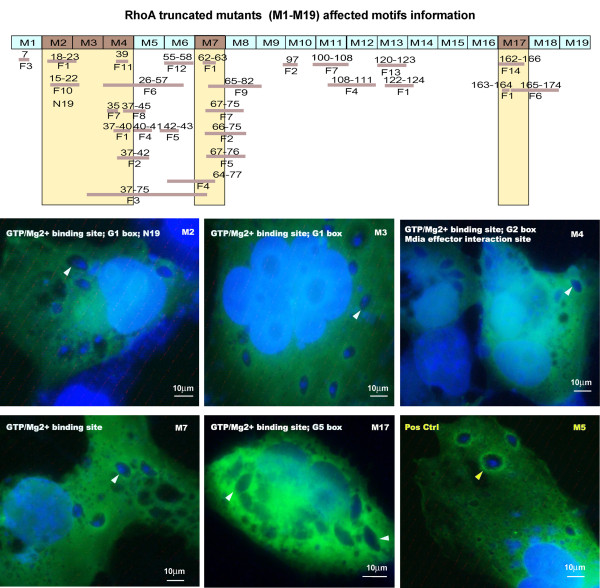
Figure 5.
The recruitment of RhoA to T. gondii PVM is dependent on different RhoA domains (1000×). COS-7 cells were transfected with 3 μg of pEGFP-N1-RhoA mutants’ plasmids M1-M19, respectively. Forty-eight hr post-transfection, the cells were infected with RH strain tachyzoites of T. gondii. M2 (RhoAΔ11–20), M3 (RhoAΔ21–30), M4 (RhoAΔ31–40), M7 (RhoAΔ61–70) and M17 (RhoAΔ161–170) were found not to accumulate on the PVM (white arrowhead and white labeling), indicating that the integrity of the features (F) as follows are essential for the recruitment of RhoA to the PVM: F1-GTP/Mg2+ binding site [chemical binding site], F-7:mDia effector interaction site, F-10:G1 box, F-11:G2 box, F-14:G5 box. The other mutants were all equally well recruited to the PVM as RhoA wild-type (yellow arrowhead in M5 is representative, and the other mutants information is provided in Additional file 3: Data S3), indicating that the other motifs of RhoA such as F2-GAP (GTPase-activating protein) interaction site [polypeptide binding site], F3-GEF (guanine nucleotide exchange factor) interaction site [polypeptide binding site], F4-GDI (guanine nucleotide dissociation inhibitor) interaction site [polypeptide binding site], F5-Rho kinase (ROCK) effector interaction site [polypeptide binding site], F6-PKN/PRK1 effector interaction site, F8-Switch I region, F9-Switch II region, F12-G3 box and F13-G4 box, are not the decisive motifs for the recruitment of RhoA to the PVM. Bar: 10 μm.