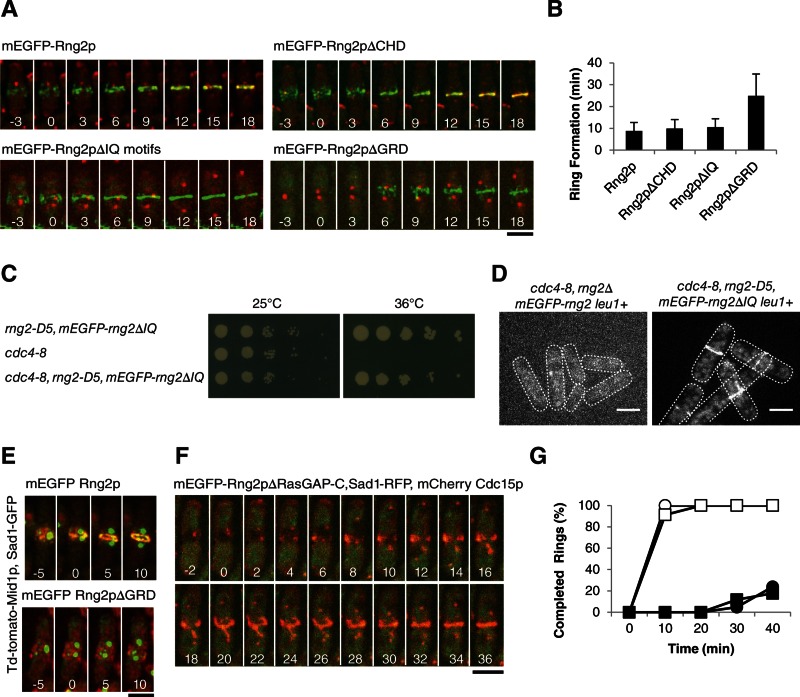
FIGURE 4:
Ability of Rng2p constructs lacking each of the first three domains to restore cytokinesis of rng2Δ cells. (A) Time series of fluorescence micrographs at 3-min intervals at 25°C of rng2Δ cells expressing Sad1-RFP (red) and complemented with full-length mEGFP-Rng2p or the indicated deletion constructs of mEGFP-Rng2p (green). mEGFP-Rng2p and mEGFP-Rng2pΔCHD cells also expressed mCherry-Cdc15p (red). Time in minutes is shown in white, with SPB separation defined as time zero throughout. (B) Average time (±1 SD) after SPB separation for each strain to form a ring using the mEGFP-Rng2p or mEGFP-Rng2p deletion construct to mark contractile rings (n = 13, 21, 8, and 27, respectively). (C) Complementation of cdc4-8 cells with mEGFP-rng2ΔIQ. (D) Fluorescence micrograph of mEGFP-Rng2p or mEGFP-Rng2pΔIQ localization in cdc4-8 cells at 32°C. (E) Time series of fluorescence micrographs at 5-min intervals at 25°C of rng2Δ cells expressing Sad1-GFP (green), Mid1p-TdTomato (red), and the indicated mEGFP-Rng2p deletion construct (green). A maximum intensity projection was formed from 13 Z-slices collected for each time point. (F) Time series of fluorescence micrographs at 2-min intervals at 32°C of rng2-D5 cells expressing Sad1-RFP (red), mCherry-Cdc15 (red), and mEGFP-Rng2pΔRasGAP-C (green). (G) Outcomes graph of percentage of cells with complete rings as a function of time after SPB separation at 32°C: (◯) rng2+ cells (n = 27); (●) rng2-D5 cells (n = 21); (□) rng2-D5, mEGFP-rng2 cells (n = 23); (■) rng2-D5, mEGFP-Rng2pΔRasGAP-C cells (n = 17). Scale bars, 5 μm.