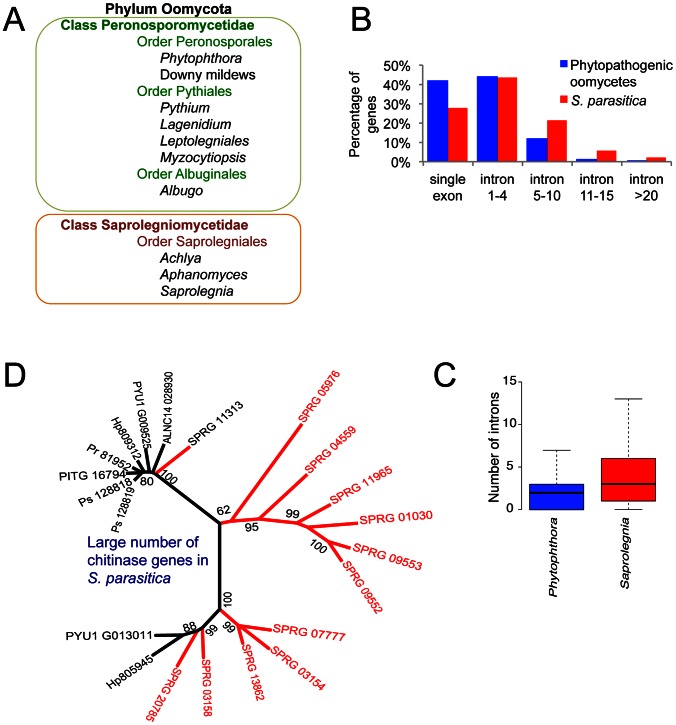
Figure 1. Taxonomy and ancestral genomic features in S. parasitica.
(A) Animal pathogenic and plant pathogenic oomycetes reside in different taxonomic units. (B) Comparison of intron number in phytopathogenic oomycetes (the average count from the total genes of P. infestans, P. ramorum, P. sojae, Py. ultimum and H. arabidopsidis) and S. parasitica among all genes. (C) Significant difference in intron number in 4008 orthologous genes shared by S. parasitica and Phytophthora species (average intron count of P. infestans, P. sojae and P. ramorum). (Wilcoxon test, p<0.001). (D) Large number of chitinase genes belonging to CAZy family GH-18 in S. parasitica (red) compared to other oomycetes (black; Ps = P. sojae, Pr = P. ramorum, PITG = P. infestans, Hp = H. arabidopsidis, Pyu = Py. ultimum, ALNC = A. laibachii). The phylogenetic tree was constructed with chitinase genes from oomycetes using Maximum likelihood method.