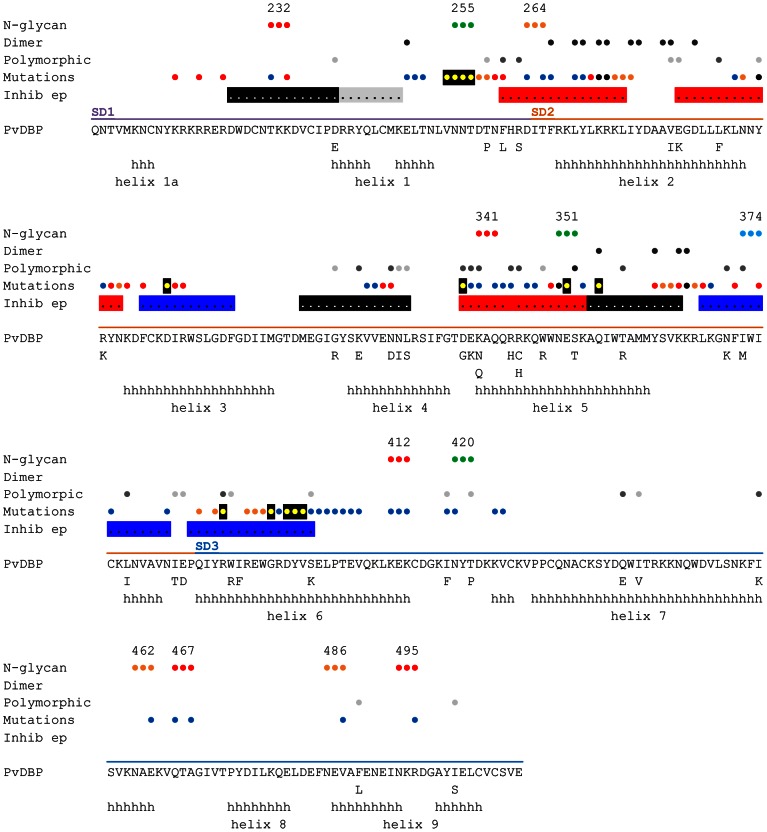
Figure 2. Summary of PvDBP polymorphism, inhibitory epitopes, and residues impacting DARC binding.
The sequence of the solved Sal1strain PvDBP variant crystal structure [13] is shown. Polymorphic amino acids from 129 PvDBP sequences [28], [29] are listed below the Sal1 sequence. Alpha helices in the PvDBP crystal structure [13] are indicated by “h” and labeled helix 1a to helix 9 according to convention [71]. Circles above the line-up indicate important residues. N-glycosylation sites are numbered according to Table 1 and colored green (wild type), blue (STBP glycan), orange (P1 and Max), and red (Max). Dimer interface [13] – black circles; polymorphism – light grey (rare, <10% of sequences), dark grey (>10% of sequences); mutations that effect DARC binding from this study and others [13], [17]–[19], are colored blue (no effect), yellow with black shadowing (minor), orange (moderate), red (major), and black, differences between studies (see Table S1); linear epitopes targeted by inhibitory antibodies [20] – black or grey shading (low inhibitory), blue shading (medium inhibitory), red shading (high inhibitory).