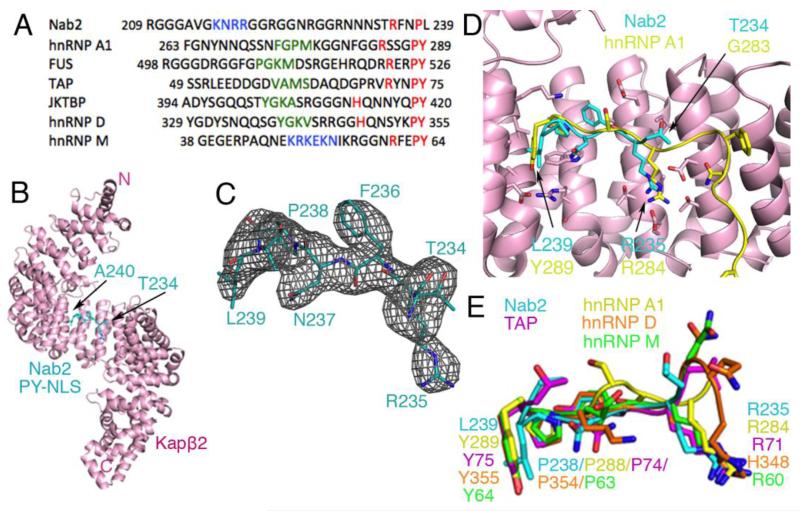
Fig. 1. Structure and comparative analysis of Nab2PY-NLS.
(A) Sequence alignment of several PY-NLSs. This signal consists of an N-terminal hydrophobic or basic motif (green and blue, respectively) and a C-terminal RX2-5PY motif (red). (B) Overall structure of the Kapβ2•Nab2PY-NLS complex. The karyopherin is in pink and the PY-NLS cyan. (C) mFo-DFc difference map, contoured at 2.0σ (grey), at the PY-NLS binding site of Kapβ2 before the Nab2PY-NLS residues were included in the model. The final refined model of Nab2PY-NLS residues Thr234 to Ala240 are in cyan. (D) Superposition of Kapβ2 residues 301-634 for Kapβ2s bound to PY-NLSs from cargos Nab2 and hnRNP A1 (PDB ID 2H4M; Cα rmsd 0.35 Å). Kapβ2 of the Nab2 complex is shown in pink. The Nab2 PY-NLS is in cyan and the hnRNP A1 PY-NLS is in yellow. (E) Similar superpositions of Kapβ2 as in D, to compare PY-NLSs from Nab2 (cyan), hnRNP A1 (yellow), hnRNP M (green), hnRNP D (orange), and TAP/NXF1 (purple).