Background: HAMLET is a broadly tumoricidal complex of partially unfolded α-lactalbumin and oleic acid whose structural and functional contributions to HAMLET remain largely undefined.
Results: The protonation state and function of the lipid are elucidated.
Conclusion: HAMLET and oleate possess unique and overlapping effects on tumor cells.
Significance: Unfolded proteins, like α-lactalbumin, form novel functional entities with deprotonated fatty acids.
Keywords: Cancer Biology, Cell Death, Lipids, Lipid Metabolism, Protein Complexes, Protein Folding, HAMLET, Oleate, Oleic Acid
Abstract
Long-chain fatty acids are internalized by receptor-mediated mechanisms or receptor-independent diffusion across cytoplasmic membranes and are utilized as nutrients, building blocks, and signaling intermediates. Here we describe how the association of long-chain fatty acids to a partially unfolded, extracellular protein can alter the presentation to target cells and cellular effects. HAMLET (human α-lactalbumin made lethal to tumor cells) is a tumoricidal complex of partially unfolded α-lactalbumin and oleic acid (OA). As OA lacks independent tumoricidal activity at concentrations equimolar to HAMLET, the contribution of the lipid has been debated. We show by natural abundance 13C NMR that the lipid in HAMLET is deprotonated and by chromatography that oleate rather than oleic acid is the relevant HAMLET constituent. Compared with HAMLET, oleate (175 μm) showed weak effects on ion fluxes and gene expression. Unlike HAMLET, which causes metabolic paralysis, fatty acid metabolites were less strongly altered. The functional overlap increased with higher oleate concentrations (500 μm). Cellular responses to OA were weak or absent, suggesting that deprotonation favors cellular interactions of fatty acids. Fatty acids may thus exert some of their essential effects on host cells when in the deprotonated state and when presented in the context of a partially unfolded protein.
Introduction
Long-chain fatty acids are essential cellular components, serving as nutrients, membrane constituents, signaling molecules, and precursors for prostaglandins and other crucial bioactive substances (1). Examples of their effects include modifications of enzymatic function, gene expression, synaptic transmission, and metabolism (2–5). Dysregulated long-chain fatty acid function is associated with numerous medical disorders, including infection, inflammation, atherosclerosis, and cancer (6–9). To fulfill these diverse functions, fatty acids engage with cell membranes, and specific fatty acids are taken up from the circulation. A variety of membrane interaction mechanisms have been characterized, but many aspects remain unclear. Fatty acids cross lipid bilayers via flip-flop mechanisms or specific protein-receptor interactions. Receptors identified in caveolae/lipid rafts (10) include fatty acid translocase/Cluster of Differentiation 36 (FAT/CD36), caveolin-1, and interacting cytosolic fatty acid-binding proteins (FABs), which bind anionic phospholipids as well as lipids modified by peroxidation (11, 12).
HAMLET2 (human alpha-lactalbumin made lethal to tumor cells) is a complex of α-lactalbumin and oleic acid (13, 14) and the first member in a new family of complexes formed from partially unfolded proteins with fatty acids as integral constituents. HAMLET displays broad anti-tumor activity in vitro with a high degree of tumor selectivity (15). Studies in patients and animal models have confirmed this selectivity and demonstrated therapeutic efficacy against several tumor types (16–20). HAMLET has broad tumoricidal activity unrelated to well defined cellular responses such as apoptosis (21–23). Essential aspects of the conserved death response to HAMLET have been defined, including the dependence on oncogenic transformation (22), proteasome inhibition (24), and nucleosome-histone binding (25). We have recently identified ion channel activation as a new, unifying mechanism of HAMLET tumoricidal effect (23). Rapid ion fluxes triggered by HAMLET were shown to initiate changes in morphology, viability, gene expression, and MAPK signaling, and especially Na+ or K+ fluxes were essential for these responses to occur.
In a screen for suitable fatty acids cofactors, C18:1, cis-monounsaturated fatty acids were identified as optimal for HAMLET formation (26), suggesting that these fatty acids may share specific structural features required both for HAMLET formation and to engage targets involved in tumor cell death. In contrast, trans-poly/monounsaturated or fully saturated fatty acids failed to form a HAMLET-like complex under near identical experimental conditions (26–29). The contribution of the C18:1 cis fatty acids to the tumoricidal activity of HAMLET has been debated, however. Studies of complexes with high lipid content have recently suggested that unfolded proteins may function solely as “lipid carriers” and that the tumoricidal response is triggered by the long-chain fatty acids alone (28, 30).
The effect of lipids on host cells is influenced by the protonation state. For example, experiments using anionic inhibitors suggested that the deprotonated form of long-chain fatty acids may be the most relevant for cellular uptake (12). The protonation state of oleic acid in HAMLET is unclear due to the phase behavior of oleic acid, which increases the apparent pKa to between 8.0 and 8.5 (31) or 9.85 (32). Oleic acid is expected to be deprotonated in HAMLET, however, as the complex is formed by ion exchange chromatography at basic pH. Oleate has been used successfully to form HAMLET-like complexes (33), but structural or biological differences between oleic acid and oleate as cofactors in HAMLET have not been examined.
To address this question, we first show by 13C NMR that oleate and oleic acid produce structurally homologous HAMLET complexes. Using a variety of cellular assays, we subsequently identify dose-dependent effects of oleate on tumor cells, partially overlapping with those of HAMLET. Fatty acids may thus exert some of their essential effects on host cells when in the deprotonated state and when presented in the context of a partially unfolded protein.
EXPERIMENTAL PROCEDURES
HAMLET/Oleate-HAMLET Production and Cell Culture
α-Lactalbumin was purified from defatted human milk by ammonium sulfate precipitation and a hydrophobic interaction column and converted to HAMLET by removal of calcium and binding to oleic acid as previously described (14). For oleate-HAMLET, oleic acid was replaced by sodium oleate (Sigma) dissolved in Tris buffer, pH 8.5, by vortexing.
Lung carcinoma cells (A549) and T-cell lymphoma cells (Jurkat) (ATCC, Manassas, VA) were cultured in RPMI 1640 with nonessential amino acids (1:100), 1 mm sodium pyruvate (all from PAA, Pasching, Austria), 50 μg/ml gentamicin (Invitrogen), and 5% fetal calf serum (FCS).
Circular Dichroism (CD) Spectroscopy
Far- and near-UV CD spectra were collected on HAMLET and oleate-HAMLET at 25 °C using a Jasco J-810 spectropolarimeter as previously described (14). Lyophilized HAMLET and oleate-HAMLET were dissolved in phosphate-buffered saline (PBS). Secondary structure predictions were made using K2D3 (34) with 41 input points from 200 to 240 nm in Δϵ units calculated as Δϵ = θm/3298. Thermal denaturation measurements at 270 and 222 nm were collected from 5 to 95 °C at a scan rate of 60 °C h−1, 1-nm wavelength step, 8-s response time. HAMLET and oleate-HAMLET were reconstituted in PBS at 32 and 43 μm, respectively, for 270 nm and to 28 μm for 222-nm measurements. Data were presented as above. The extent of unfolding was calculated by taking the ratio of the measured ellipticity at each temperature to the maximum ellipticity at 270 nm.
Nuclear Magnetic Resonance (NMR) Spectroscopy
1H and natural abundance 13C NMR experiments were performed on an 800-MHz (18.8 tesla) Premium COMPACT 54 mm/UHF DD2 NMR spectrometer with HCN Cold Probe (Agilent Technologies). Lyophilized samples were dissolved in either 100% D2O (or 80%/20% H2O/D2O), 50 mm sodium phosphate buffer, pH 7.4, with 1,4-dioxane added to serve as chemical shift standard (3.75 ppm for 1H, 67.3 ppm for 13C). Oleic acid and oleate (both from Sigma) were dissolved in 100% methanol for determination of the chemical shifts of protonated and deprotonated carboxyl carbons. The following parameters were used for the 13C NMR acquisition: temperature 25.0 °C, 90-degree pulse width 14.5 μs, acquisition time 0.164 s, spectral width 50000 Hz (−20 to 220 ppm), relaxation delay 9.0 s, data points 16,000, and scans 8000.
Cell Death Analysis
A549 lung carcinoma cells (5 × 105 cells/ml in suspension) were treated with HAMLET, oleate-HAMLET, oleic acid, or sodium oleate at concentrations described under “Results,” and cell death was measured by the reduction in ATP level and trypan blue exclusion as previously described (23) and Prestoblue cell viability reagent (Invitrogen) according to manufacturer's instructions. Oleic acid was dissolved in ethanol and diluted to 3.5 mm in PBS and sonicated. Sodium oleate was dissolved directly in PBS to 3.5 mm by vortexing and sonication.
Phase Holographic Imaging
The HoloMonitorTM M3 digital holographic microscope (Phase Holographic Imaging AB, Lund, Sweden) records three-dimensional information using interfering wave fronts induced by the exposure to a 0.8-milliwatt HeNe laser (633 nm) (35–37). The interference pattern (hologram) is recorded on a digital sensor and is used to reconstruct the amplitude and phase of the object (38, 39). 40,000 A549 cells were cultured on μ-Slide I coated with ibiTreat (ibidi, Martinsried, Germany) overnight. The cells were washed and replaced with fresh serum-free RPMI medium and exposed to 35 mm HAMLET, 175 mm oleate or oleic acid for 3 h at 37 °C, 5% CO2. 5% FCS was added after 1 h of incubation. The holograms were captured with an imaging time of 2.4 ms every 15-min interval for the first hour and at 90, 120, and 180 min after treatment.
Intracellular Ion Concentrations and Ion Fluxes
The relative, free intracellular concentrations of Ca2+ ([Ca2+]i) and Na+ ([Na+]i) were estimated using the calcium indicator Fluo4 NW and the sodium fluorophore CoroNa Green, respectively (Invitrogen), as previously described (23). For estimation of K+ fluxes, the FluxOR™ potassium ion channel assay (Invitrogen) was used according to the manufacturer's instructions. Briefly, cells were incubated with FluxOR™, which is a Tl+ indicator. An increase in fluorescence signal, which was measured at 535 nm after excitation at 485 nm, corresponds to an influx of Tl+, indicating opening of potassium channels.
Confocal Microscopy
A549 lung carcinoma cells were seeded overnight at 37 °C, 5% CO2 on 8-well chamber slides (Nalge Nunc, Rochester, NY). The cells were washed twice with serum-free RPMI medium and incubated with HAMLET, oleate-HAMLET, oleic acid, or sodium oleate at equimolar concentrations for a period of 30 min to 3 h in RPMI medium at 37 °C, 5% CO2. 5% FCS were added after 1 h of incubation. The cells were fixed with 3.7% formaldehyde. The supernatant with detached cells was fixed with formaldehyde separately and cytospun at 500 rpm for 5 min onto l-lysine-coated glass slides (Thermo Scientific) for analysis. The cells were stained with 100 μl of Nile Red (9-diethylamino-5H-benzo[α]-phenoxazin-5-one, 100 μg/ml) (Sigma) for 5 min at room temperature. Nuclei were strained using DRAQ5 (eBioscience, San Diego, CA).
The slides were analyzed with LSM 510 META system (Carl Zeiss, Jena, Germany) with two spectra settings; excitation with 488 nm and emission analysis with a band pass 505–550 filter for yellow-gold fluorescence and excitation with 543 nm and emission analysis with a long pass 585 filter for red fluorescence.
Transcriptomics
A549 cells (200 000/well) were allowed to adhere overnight on a 6-well plate (TPP, Trasadingen, Switzerland). After exposure to HAMLET (1 h, 35 μm), oleic acid, or sodium oleate (both from Sigma, 1 h, 175 μm), RNA was extracted (RNeasy Mini kit, Qiagen). The samples were analyzed using standard Affymetrix protocols. The raw data were normalized using RMA (40) as provided by R and Bioconductor. Probe sets with a p value < 0.05 and log 2-fold change >1 were considered differentially expressed. To functionally characterize the resulting gene lists, Database for Annotation, Visualization, and Integrated Discovery (41) and Ingenuity Pathway Analysis (Ingenuity Systems®) were employed.
Real-Time PCR
Total RNA was reverse-transcribed using SuperScriptTM III First-Strand Synthesis System for RT-PCR (Invitrogen) and quantified in real time using QuantiTect SYBR Green based PCR kits for mRNA (Qiagen) on a Rotor Gene Q (Qiagen). Quantitative real time-PCR reactions were run in biological duplicates and triplicates, and gene expression was done based on the comparison with GAPDH expression.
Metabolomics
HAMLET-treated cells were washed with phosphate-buffered saline followed by the addition of ice-cold methanol to stop metabolic activity. Subsequently, cells were scraped into the solvent and transferred to a reaction tube. Six replicates consisting of three pooled wells each were collected per sample group. For extraction, the methanol phase was removed from the cells, dried, and stored at −80 °C. Cell pellets were dried in a FreeZone 2.5 lyophilizer (Labconco, Kansas City, MO), homogenized using a Mini-Beadbeater (BioSpec Products, Bartlesville, OK), and extracted a pre-cooled methanol-isopropyl alcohol-water (3:3:2) mixture. A mixture of internal retention index markers was prepared using fatty acid methyl esters of C8, C9, C10, C12, C14, C16, C18, C20, C22, C24, C26, C28, and C30 linear chain length dissolved in chloroform at a concentration of 0.8 mg/ml (C8-C16) and 0.4 mg/ml (C18-C30). 1 ml of this retention index mixture was added to the dried extracts. 10 μl of a solution of 40 mg/ml 98% pure methoxyamine hydrochloride (Chemical Abstracts Service no. 593-56-6, Sigma) in pyridine (silylation grade, Pierce) was added and shaken at 30 °C for 90 min to protect aldehyde and ketone groups. 90 μl of N-methyl-(trimethylsilyl) trifluoroacetamide and 1% trimethylchlorosilane (1 ml bottles, Pierce) was added for trimethylsilylation of acidic protons and shaken at 37 °C for 30 min. A Gerstel (Mülheim an der Ruhr, Germany) automatic liner exchange system with a multipurpose sample MPS2 dual rail was used to inject 0.5 ml of sample into a Gerstel CIS cold injection system (Gerstel). Samples were injected into a 50 °C injector port and held for 1 min. They were then separated using an Agilent 6890 gas chromatograph equipped with a 30-m-long, 0.25-mm inner diameter Rtx5Sil-MS column (Restek (Bellefonte, PA), 0.25-mm 5% diphenyl film, and additional 10-m integrated guard column) by ramping to 330 °C at 20 °C min−1 and holding for 5 min. Mass spectrometry was performed on a Leco Pegasus IV time-of-flight mass spectrometer (St. Joseph, MI) with a 280 °C transfer line temperature, −70-eV electron ionization, and an ion source temperature of 250 °C. Mass spectra were acquired from m/z 85 to 500 at 17 spectra/s and 1850-V detection. Result files were processed using the metabolomics BinBase database. All database entries in BinBase were matched against the Fiehn mass spectral library of B1200 authentic metabolite spectra using retention index and mass spectrum information or the NIST05 commercial library. Identified metabolites were included if present within at least 50% of the samples of each treatment group (as defined in the SetupX database).
RESULTS
Oleate Is the Functional Cofactor in HAMLET
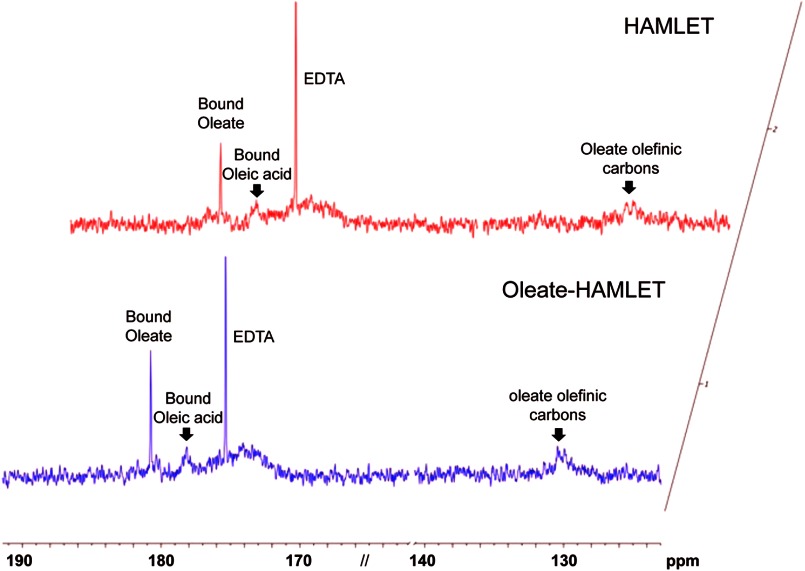
The protonation state of bound oleic acid in HAMLET was determined by natural abundance 13C NMR spectroscopy. Protonated and deprotonated carboxyl groups were clearly distinguishable from the overlay of the two spectra (supplemental Fig. S1A). Carboxyl carbon of oleate showed a chemical shift at 182 ppm, whereas that of oleic acid was 177 ppm. Both sodium oleate and oleic acid dissolved in methanol or sodium phosphate buffer gave rise to a 130 ppm peak, which corresponds to the olefinic carbons. Peaks further upfield correspond to the aliphatic carbons. The natural abundance of 13C in HAMLET was determined (Fig. 1). HAMLET showed a prominent peak at 182 ppm and a broad 130-ppm peak, a clear difference from free fatty acid. A minor bound oleic acid peak was also detected. An additional peak at 175 ppm, verified to be the carboxyl carbon of residual EDTA in the conversion process, was also recorded. These results identify oleate as the lipid cofactor in the HAMLET complex.
FIGURE 1.
Natural abundance 13C NMR spectra of HAMLET (upper panel) and oleate-HAMLET (lower panel). Carboxyl and olefinic carbon regions of 13C NMR in 100% D2O (or 80%/20% H2O/ D2O), 50 mm sodium phosphate buffer, pH 7.4. Chemical shift positions of bound oleate/oleic acid, residual EDTA, and olefinic carbons are marked. HAMLET and oleate-HAMLET showed an identical prominent peak at 182 ppm and a broad 130 ppm peak, indicating that oleate is bound to both complexes.
Oleate and Oleic Acid Form Homologous HAMLET Complexes
To confirm the protonation state of oleic acid in HAMLET, ion exchange matrices were pre-conditioned with oleic acid or sodium oleate, as described (14). The preconditioned columns were loaded with EDTA-treated, partially unfolded human α-lactalbumin and eluted using a NaCl gradient. HAMLET and oleate-HAMLET eluted as sharp peaks at 0.7 m NaCl (supplemental Fig. S1, A and B). By near-UV CD spectroscopy (250–320 nm wavelength) (supplemental Fig. S1C), HAMLET and oleate-HAMLET showed a similar decrease in signal intensity at 270 nm as compared with native human α-lactalbumin (14). By far-UV CD spectroscopy, both complexes were shown to retain similar secondary structure contents (supplemental Fig. S1D) (14). Further structural characterization by thermal denaturation followed by CD spectroscopy confirmed the structural similarity (supplemental Fig. S1, E–G), but unfolding of HAMLET occurred at a lower temperature. The melting temperature, Tm, estimated from the thermal denaturation profiles for native α-lactalbumin and HAMLET, were ∼60 °C and ∼31 °C, respectively (42, 43), and the recorded Tm oleate-HAMLET was ∼35 °C (supplemental Fig. S1E). Natural abundance 13C NMR of oleate-HAMLET confirmed that oleate is the bound state in both complexes and shows a prominent peak at 182 ppm and a broad 130-ppm peak (Fig. 1). HAMLET and oleate-HAMLET thus show a high degree of structural similarity, including almost identical elution profiles, similar secondary and tertiary structure characteristics, and melting temperature.
Tumor Cell Death in Response to HAMLET, Oleate-HAMLET, Oleate, and Oleic Acid
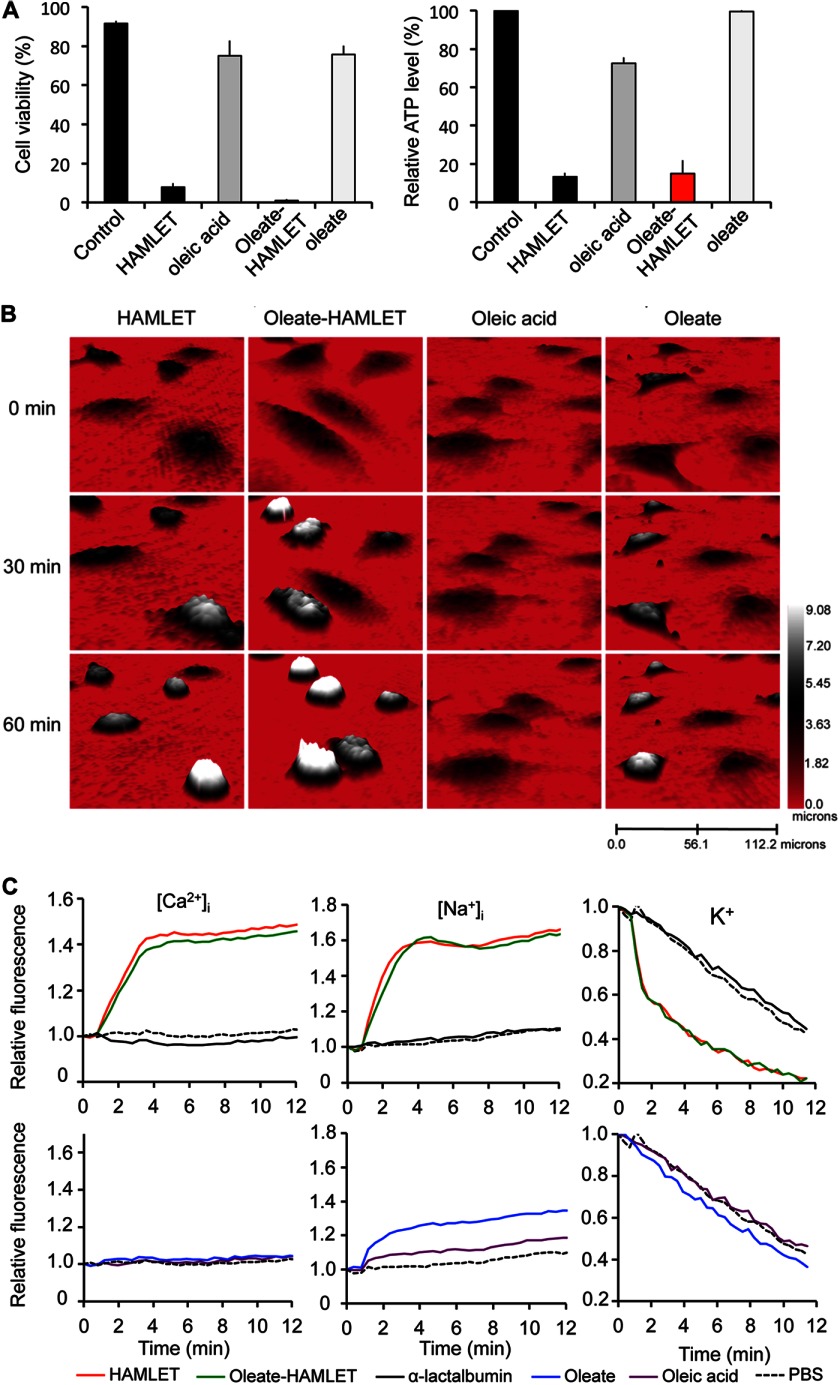
To ensure that oleate-HAMLET reproduces the tumoricidal effect of HAMLET, cell death was quantified in A549 lung carcinoma cells exposed to the respective complex (Fig. 2A). No difference in dose-dependent cytotoxicity was recorded between HAMLET and oleate-HAMLET (>80% dead cells after 3 h at 35 μm) (supplemental Fig. S2A).
FIGURE 2.
HAMLET and oleate-HAMLET trigger tumor cell death, morphological change, and ion fluxes. A, cytotoxic responses in A549 lung carcinoma cells were treated with HAMLET or oleate-HAMLET (35 μm) and quantified by trypan blue exclusion and ATP levels. Sodium oleate- or oleic acid (175 μm) were not active at the molar concentration present in HAMLET/oleate-HAMLET. B, morphological response to HAMLET, oleate-HAMLET (1 h, 35 μm), sodium-oleate or oleic acid (1 h, 175 μm) was recorded by holographic imaging. Rounding up of cells was observed after HAMLET and oleate-HAMLET treatment, and oleate caused a partial effect. C, the relative free, intracellular concentrations of Na+, K+, or Ca2+ ([Na+]i, [K+]i, and [Ca2+]i) were estimated in A549 cells (K+ and Ca2+) or Jurkat cells (Na+) by fluorescence spectrometry using CoroNa Green, FluxOR, or Fluo-4. Compared with HAMLET (35 μm), oleate (175 μm) caused a moderate efflux of potassium and no influx of calcium or sodium in A549 or Jurkat cells, respectively.
The lipid concentration in the HAMLET/oleate-HAMLET complexes was determined as ∼1:4 or 1:5 by acid hydrolysis and GC/MS (26). At a concentration corresponding to five oleate molecules (175 μm) per protein (35 μm α-lactalbumin), oleate or oleic acid did not alter the viability of lung carcinoma cells (3 h) compared with control (not significant, supplemental Fig. S2B). At higher lipid concentrations (15 times), oleate was more efficient as a cytotoxic agent than oleic acid (supplemental Fig. S3B). At 25 times, all cells underwent rapid lysis, confirming well known effects of high lipid concentrations on cellular integrity.
Effects on tumor cell morphology were compared by confocal microscopy (supplemental Fig. S2C). In response to HAMLET and oleate-HAMLET (35 μm, 1 h), the cells rounded up and detached. By real time holography imaging (Fig. 2B), HAMLET and oleate-HAMLET caused a parallel reduction in cell surface area and an increase in height. Less pronounced morphological changes were observed in cells treated with oleate alone, and oleic acid left the cells largely unchanged. By transmission light microscopy, all treated cells showed an increase in granularity indicative of lipid droplet formation, suggesting uptake of both the protonated and deprotonated fatty acid (supplemental Fig. S2C).
Differences in Ion Channel Activation by HAMLET and Oleate/Oleic Acid
HAMLET triggers ion fluxes across cell membranes and ion channel inhibitors; blocking such fluxes prevents HAMLET uptake as well as tumor cell death (23). To address if oleate is an independent trigger of ion fluxes, ion channel activation was quantified in lung carcinoma or Jurkat cells preloaded with Ca2+ or Na+ fluorophores or thallium, a surrogate marker for K+. HAMLET and oleate-HAMLET triggered rapid Na+, K+, and Ca2+ fluxes of similar magnitude (Fig. 2C). A weak Na+ and K+ response to oleate was recorded, but no Ca2+ response was detected. Increasing the oleate concentration to a cytotoxic level of 500 μm did not entirely reproduce the ion flux patterns induced by HAMLET (supplemental Fig. S2D).
The Metabolic Response to Oleate
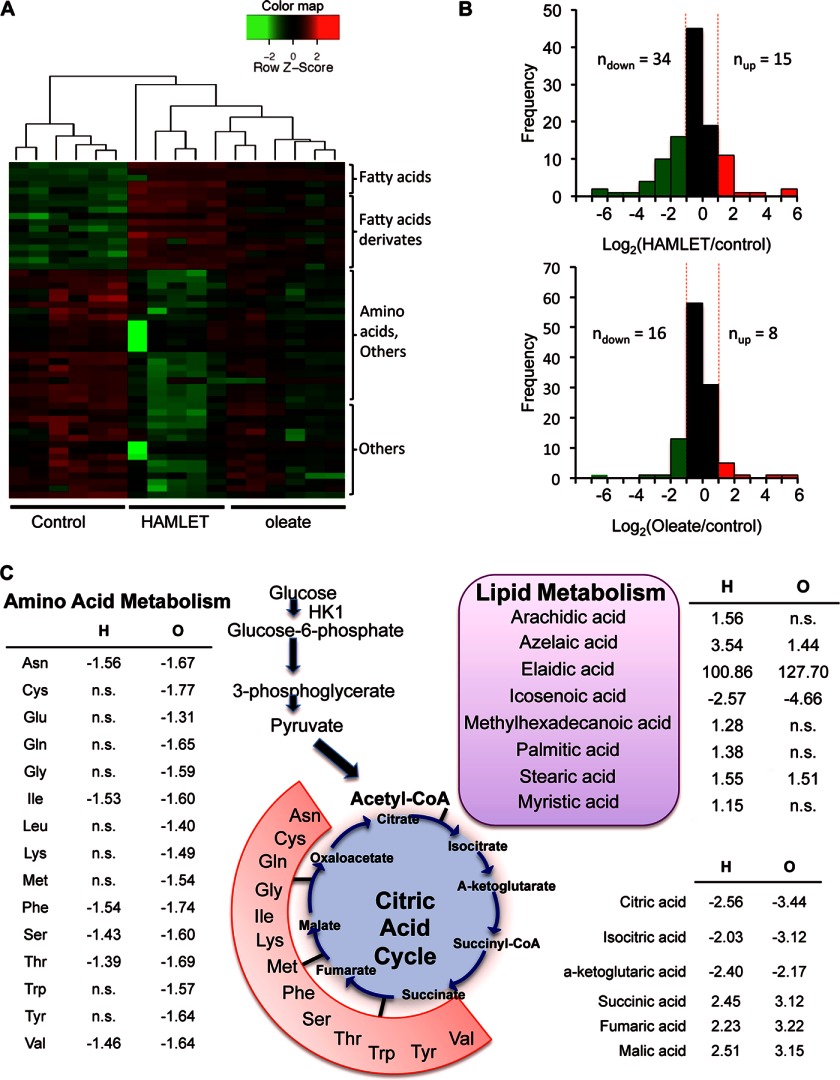
HAMLET causes metabolic paralysis in tumor cells, with a reduction >60% in abundance of metabolites after 1 h (22). To examine the effects of oleate on the metabolic response, oleate-treated cells (60 min, 175 μm) were subjected to a non-targeted metabolite profiling by gas chromatography-mass spectrometry (Fig. 3A). After HAMLET treatment, 34 of the 49 altered metabolites were reduced compared with 16 of 24 in oleate-treated cells (Fig. 3B). An increase in oleic acid, elaidic acid, and icosenoic acid was detected, possibly reflecting an increase in fatty acid uptake by the cells. Decreased metabolites after oleate treatment included pyrophosphate, aconitic acid, glycine, taurine, and Tris (supplemental Fig. S3). In contrast, only three metabolites were altered by oleic acid (60 min, 175 μm), with an increase in pentadecanoic acid and tyrosine (3.6- and 3.8-fold) and decrease in elaidic acid compared with control (4-fold) (supplemental Fig. S3). The results suggested major differences in the effects of HAMLET and oleate on tumor cell metabolism.
FIGURE 3.
Metabolic responses to HAMLET, oleate, or oleic acid. Metabolites were quantified by GC/MS in extracts of A549 cells treated with HAMLET (35 μm) or oleate (175 μm) for 60 min. A, shown is a heatmap of differentially abundant metabolites. A global decrease in metabolite abundance was detected in the HAMLET samples. B, frequency distribution of log 2-transformed-fold changes in signal intensity between HAMLET-treated and oleate-treated cells is shown. C, targeted metabolite profiling is shown. Change in abundance of lipids, amino acids, and the citric acid cycle metabolites in HAMLET- or oleate-treated cells is shown. n.s., not significant.
To further characterize the metabolic response, HAMLET- or oleate-treated cells were subjected to targeted metabolite profiling of fatty acids, amino acids, and citric acid cycle constituents (Fig. 3C). In total, 43 metabolites were significantly altered by HAMLET treatment (p value < 0.05, supplemental Table S1). Consistent with the non-targeted profiling, an increase in fatty acid metabolites was detected (stearic, palmitic, methylhexadecanoic, elaidic, azelaic, arachidic, and myristic acid; Fig. 3C). There was a loss of early metabolites in the citric acid cycle (citric, isocitric, and α-ketoglutaric acid) and an accumulation of late metabolites (succinic, fumaric, and malic acid). Furthermore, metabolites feeding into the citric acid cycle were reduced, in particular, sorbitol, fructose, and glycerol-α-phosphate and amino acids (asparagine, isoleucine, phenylalanine, serine, threonine, and valine). In parallel, a late glycolytic intermediate (3-phosphoglycerate) and a carbohydrate metabolism derivative (5-hydroxy-methyl-2-furoic acid) were increased after HAMLET treatment.
In the targeted metabolic profiling, the effects of oleate were at least as pronounced as for HAMLET. In total, 61 metabolites were significantly altered (p value < 0.05; supplemental Table S1). Fatty acid metabolites were less strongly affected than by HAMLET (azelaic, elaidic, and stearic acid were increased), and the loss of early and accumulation of late metabolites in the citric acid cycle was similar to HAMLET. The deregulation of amino acid metabolism was more apparent in oleate- than in HAMLET-treated cells, with an additional nine decreased amino acids or amino acid derivatives.
The results suggest that HAMLET and oleate have similar effects on the TCA cycle but that the pronounced effects of HAMLET on overall tumor cell metabolism are not reproduced by oleate (175 μm). The general decrease in amino acids in HAMLET and oleate-treated cells might indicate that amino acids are used as an alternative source of energy when the proximal TCA cycle is blocked.
Effects of HAMLET and Oleate/Oleic Acid on Gene Expression
Genome wide transcriptomic analysis was used to further compare the cellular effects of HAMLET and oleate. RNA samples from lung carcinoma cells treated with HAMLET (35 μm) or oleic acid or oleate (175 μm) were hybridized to Affymetrix Whole Genome Microarrays U219. The resulting hybridization profiles were assessed pre- and post-RNA normalization. For statistical analysis, normalized data were linear model-fitted, and p values were estimated using an empirical Bayes approach. Genes with a log 2-fold change >1 and p value <0.05 were considered differentially expressed. The transcriptional response to oleic acid was restricted to two genes with a log 2-fold change >1 and p value <0.05, EGR1 and PKD4.
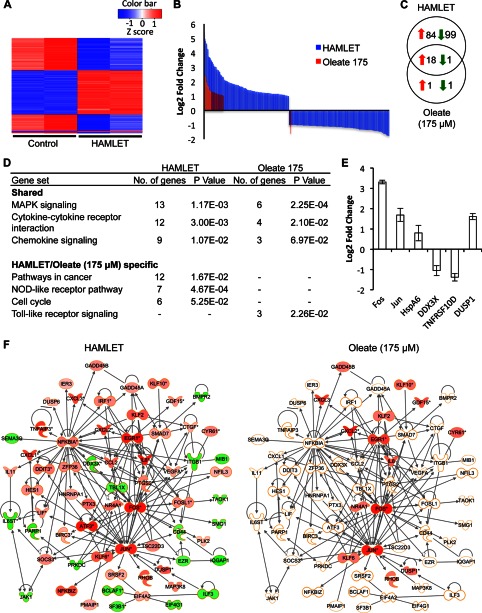
A heatmap of the top 200 differentially expressed genes by -fold change is shown in Fig. 4A. A pronounced effect of HAMLET on gene expression was observed (74 up-regulated and 128 suppressed genes) compared with oleate (19 up-regulated and 2 suppressed genes), 19 of which overlapped with HAMLET (Fig. 4, B and C). The suppressed genes (Fig. 4B) include genes associated with DNA damage and repair and cell cycle regulation (enrichment scores of 2.37 and 1.8, respectively). Increased transcription of genes associated with cell death and transcriptional regulation was recorded (enrichment scores of 4.99 and 4.68, respectively).
FIGURE 4.
Effects of HAMLET or oleate on gene expression. 549 lung carcinoma cells were treated with HAMLET (35 μm) or oleate (175 μm), and RNA was harvested after 1 h. A, shown is a heatmap of the top 200 differentially expressed genes (log 2-fold change >1 and p value < 0.05) by -fold change. B, up- and down-regulated genes in HAMLET- versus oleate-treated samples are shown. More genes were differentially regulated by HAMLET than by oleate. C, HAMLET caused differential expression of 202 genes, 19 of which were shared by oleate. D, shown are shared or specific canonical pathways regulated by HAMLET or oleate. E, confirmation of gene expressions for FOS, JUN, HSPA6, DDX3X, TNFRSF10D, and DUSP1 using RT-PCR. F, a network of cancer-related genes differentially regulated by HAMLET is shown. Down-regulated genes were colored in green, and up-regulated genes are in red. Expression values from the oleate sample were overlaid onto the same network, showing that most genes were not differentially regulated in the oleate sample.
Top canonical pathways affected by HAMLET included the MAPK signaling pathway, cytokine-cytokine receptor interactions, and chemokine- and NOD-like receptor signaling pathway as well as genes in Pathways in cancer (Fig. 4D). Four p38 regulators were up-regulated including HSPA6 (2.8-fold) and three dual specificity phosphatases (DUSP1, DUSP5, and DUSP6, 2.4-, 2.1-, and 1.1-fold, respectively) acting as feedback regulators of MAPK signaling. Transcription factors that respond to DNA damage and ER stress were up-regulated (GADD45A/B and DDIT3/4, 1.5-, 1.8-, 1.8-, and 1.9-fold) as were genes implicated in cell death, including ATF3, MAP4K5, and BIRC3. Transcription factor analysis identified 37 NFκB-regulated transcripts (z-score 4.72 for NFκB) including FOS, ATF3, IL8, EGRR1, CCL20, and JUN (all with a log 2-fold change >4). The 27 p53-dependent transcripts (z-score 2.85 for p53) included PHLDA1, GADD45A, DDIT3, and DDIT4. Ten histone deacetylase-regulated transcripts were suppressed (Z-score −2.94, for histone deacetylase), consistent with previously published effects of HAMLET on histone acetylation (44).
Networks associated with cancer, cellular movement and morphology, cell cycle, cellular assembly, and organization were either exclusively found in the HAMLET-treated cells or had a lower score in the oleate (175 μm) sample (Fig. 4F). Pathways overlapping between HAMLET and oleate included cytokine-cytokine receptor interactions and the MAPK-, chemokine-, and NOD-like receptor signaling pathway, although the signals were weaker in oleate-treated cells (Fig. 4D).
The 183 HAMLET-specific genes were associated with transcription (39 genes), intracellular signaling cascades (25 genes), phosphate metabolic processes (20 genes), and cell cycle (21 genes). Ingenuity Pathway Analysis identified 131 cancer-related genes, half of which formed a cancer-related network including NFκB1A, MAP3K8, SMAD7, and DUSP6 (Fig. 4F). Two oleate-specific genes were CUL3, involved in polyubiquitination and endosome maturation, and ANGPTL4, which acts as a regulator of angiogenesis.
Differential gene expressions of FOS, JUN, TNFRSF10D, and DUSP1 for oleate (175 μm) were confirmed by RT-PCR (Fig. 4E). Two additional genes that were not found to be differentially expressed in the microarray, HSPA6 and DDX3X, were also included and validated, as their expression levels were less than the predetermined cut-off expression value.
The results suggest that most HAMLET effects on gene expression and signaling are not reproduced by oleate. Minor overlap between oleate and HAMLET-induced responses was observed.
Transcriptional Response to Higher Oleate Concentrations
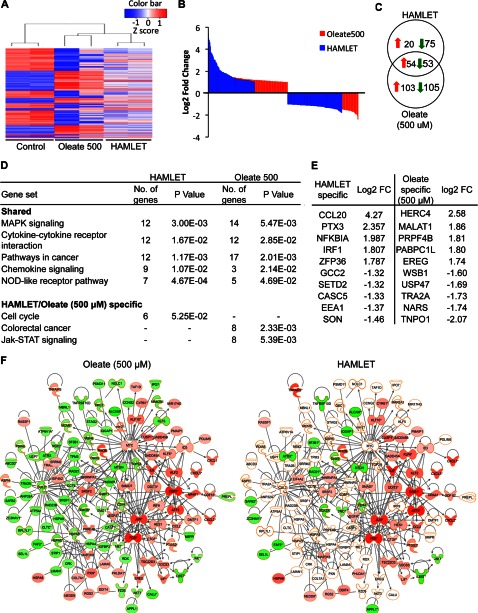
To address if a higher lipid concentration might reproduce the effects of HAMLET more fully, tumor cells were treated with 500 μm oleate. A heatmap of significantly regulated genes is shown in Fig. 5A. The higher oleate concentration triggered a significant response (157 up-regulated and 158 suppressed genes) showing more overlap with the HAMLET sample (107/202 genes, Fig. 5, B and C). Overlapping pathways included MAPK signaling, cytokine, and chemokine signaling pathways (Fig. 5D). A significant overlap was also seen in broad cancer-related networks, where the high oleate sample stimulated a larger number of genes than HAMLET.
FIGURE 5.
Gene expression in response to HAMLET or oleate (500 μm). 549 lung carcinoma cells were treated with HAMLET (35 μm) or oleate (500 μm), and RNA was harvested after 1 h. A, shown is a heatmap of the top 200 differentially expressed genes (log 2-fold change >1 and p value < 0.05). B, shown are up- and down-regulated genes for HAMLET and oleate 500 samples. More genes were differentially regulated for oleate 500 compared with HAMLET. C, a Venn diagram shows unique and overlapping genes in HAMLET or oleate 500 samples. D, shown are shared or specific canonical pathways regulated by HAMLET or oleate. E, top genes unique to HAMLET and oleate 500 are shown. F, shown is a network of cancer-related genes differentially regulated by oleate 500. Expression values from the oleate sample were overlaid onto the same network, showing that most genes were not differentially regulated in the HAMLET sample. Down-regulated genes were colored in green, and up-regulated genes are in red.
By Database for Annotation, Visualization, and Integrated Discovery (DAVID), genes specific for the high oleate sample were shown to be associated with RNA processing (16 genes), intracellular transport (13), cellular macromolecule catabolic processes (15), cell cycle regulation (18), and RNA splicing (12). By Ingenuity Pathway Analysis, 54 oleate-specific, cancer-related genes were identified, half of which formed a cancer-related network including MYC, KRAS, PXN, and MET (Fig. 5F). After exclusion of genes shared with the high oleate samples, genes that remained unique to HAMLET were mainly involved in intracellular signaling cascades (15 genes), regulation of biosynthetic process (12), and phosphate metabolic processes (12 genes). Networks involved in the maintenance or cell death and survival were also regulated.
The results suggest that important effects on gene expression are shared between HAMLET and oleate and that the extent of overlap between HAMLET and oleate-induced genes increased with the oleate concentration. Even at high oleate concentrations, however, the majority of oleate-induced genes were not regulated in HAMLET-treated cells.
DISCUSSION
Oleic acid is a key constituent of the HAMLET complex, but the extent to which the lipid contributes to tumor cell death has remained unclear as have the mechanisms involved. HAMLET is optimally formed from 18-carbon, monounsaturated, cis-fatty acids and partially unfolded α-lactalbumin, in contrast to unsaturated trans- or saturated fatty acids, which fail to form complexes with the protein. In this study we show that the deprotonated form of oleic acid (oleate) is an active lipid cofactor in HAMLET formation and that when presented in the context of partially unfolded α-lactalbumin, oleate acts directly on the cell death machinery through membrane sensing, uptake, and metabolic processing. The lipid per se did not reproduce the cell death response to HAMLET or the metabolic paralysis, however, confirming that the protein and lipid are both required for the full-fledged tumoricidal response to HAMLET. At higher oleate concentrations, a larger part of the HAMLET profile was reproduced, but major differences in cellular response profiles were still present. Oleate recognition by tumor cells is thus quantitatively and qualitatively altered in the context of partially unfolded α-lactalbumin.
A number of HAMLET-related complexes have been prepared using different α-lactalbumin variants (45, 46), aggregation states (27), related proteins like equine lysozyme (47), bovine β-lactoglobulin, or the α isoform of pike parvalbumin (28). Some of these complexes, which contain a high number of oleic acid molecules (27), have been proposed to permeabilize cells by virtue of their fatty acid content alone, giving rise to suggestions that the protein acts merely as a fatty acid delivery vehicle (48–50). It may be speculated that upon interaction of tumor cell membranes with several HAMLET molecules, membrane patches with higher lipid concentrations might be formed, resulting in membrane disruption and necrosis. The present study clearly demonstrates that HAMLET and oleate-HAMLET complexes carry a limited number of lipid moieties and that distinct cellular responses including ion fluxes and specific death pathways are activated, arguing against nonspecific membrane disruptions as a mechanism of tumor cell death. Although we show that the lipid is active in the context of the partially unfolded protein, we found no evidence of independent cytotoxic activity at concentrations equimolar to those present in HAMLET. At higher concentrations, the lipid was cytotoxic on its own, however. Importantly, cellular response profiles at either lipid concentration were not those involved in necrosis, as would have been expected if HAMLET merely permeabilizes the cell membrane due to its lipid content.
In this study, HAMLET and oleate were shown to trigger overlapping but also fundamentally different responses in tumor cells. At equimolar concentrations, oleate alone was shown to activate Na+ fluxes but not the full ion channel repertoire seen in HAMLET-treated cells. This limited effect may explain the low transcriptional response to oleate (175 μm) and its lack of cytotoxicity. At higher oleate concentrations, effects on ion fluxes were stronger, compatible with the transcriptional changes, and the cytotoxic response and overlapping effects on transcription included cancer-related pathways, MAPKs genes, and innate immune response genes. Ion fluxes were not identical to HAMLET, however, as oleate (500 μm) caused partial activation of Na+ and Ca2+ compatible with the differences in transcriptomic profiles. The results emphasize that the HAMLET complex is functionally defined in part by oleate but possesses unique additional properties not reproduced by the lipid.
Cancer metabolism is optimized for cell growth and survival under conditions of metabolic stress (51). In addition to the shift in glycolysis known as the Warburg effect, there is evidence that fatty acid oxidation contributes to the metabolic changes accompanying oncogene overexpression (52, 53). We detected two major alterations in cellular metabolism, one entailing a global metabolic paralysis, which was seen exclusively in HAMLET-treated cells, and the second, a shift in lipid metabolites, citric acid cycle constituents, and amino acids, seen both in HAMLET and oleate-treated cells but not in oleic acid-treated cells. HAMLET and oleate both increased fatty acid metabolism, consistent with lipid recognition and uptake by tumor cells, but oleic acid had no significant effect. The inertia of cells to the protonated acid was surprising but may indicate that the de-protonated fatty acid is the more active cellular agonist, as indicated by previous, cellular uptake studies (54).
Critical starting metabolites in the citric acid cycle were less abundant after HAMLET treatment, consistent with a loss of pyruvate and inhibition of glycolysis. Previously, HK1 and the glycolytic machinery have been shown to influence HAMLET sensitivity, and direct binding of HAMLET to the kinase has been detected (22). In parallel, an increased rate of amino acid catabolism and a buildup of metabolites toward the end of the cycle were observed, further suggesting that HAMLET may force tumor cells to use glucose-independent metabolic pathways and amino acid catabolism as an energy source potentially compensating for the inhibition of glycolysis. Oleate alone seemed to direct metabolism toward amino acid catabolism, suggesting that the deprotonated lipid deregulates the TCA cycle but does not trigger death. Amino acid metabolism is important for cancer cell survival, and the use of amino acid metabolites for energy production has been proposed to offer a glucose-independent solution to ATP generation in tumor cells (55, 56).
This study resolves for the first time the question of the lipid contribution to the tumoricidal activity of HAMLET. The results confirm that the lipid is a necessary constituent to achieve the tumoricidal effect of the complex (14). This was previously inferred from the finding that partially unfolded α-lactalbumin protein alone does not trigger tumor cell death (57, 58). On the other hand, oleic acid alone was largely inactive, presenting somewhat of a paradox. In this study the deprotonated lipid is shown to be a fully functional cellular agonist both alone and in the context of α-lactalbumin. The oleate effect was concentration-dependent, as higher oleate concentrations reproduced more of the cellular response to HAMLET. However, important qualitative differences were observed between the HAMLET complex and oleate alone at equimolar or higher concentrations. Although the effects on gene expression were overlapping, significant sets of genes were uniquely responsive to oleate or HAMLET, respectively. We conclude that partially unfolded α-lactalbumin is a suitable partner for oleate to offer tumor cells the “kiss of death.”
This work was supported, in whole or in part, by National Institutes of Health Grant U54 CA 112970. This study was also supported by a Sharon D. Lund Foundation grant and by the American Cancer Society, the Swedish Cancer Society, the Medical Faculty (Lund University), the Söderberg Foundation, the Segerfalk Foundation, the Anna-Lisa and Sven-Erik Lundgren Foundation for Medical Research, the Knut and Alice Wallenberg Foundation, the Lund City Jubileumsfond, the John and Augusta Persson Foundation for Medical Research, the Maggie Stephens Foundation, the Gunnar Nilsson Cancer Foundation, the Inga-Britt and Arne Lundberg Foundation, and the HJ Forssman Foundation for Medical Research and the Royal Physiographic Society. Support was also obtained from the Danish Council for Independent Research (Medical Sciences).

This article contains supplemental Figs. S1—S3 and Table S1.
- HAMLET
- human α-lactalbumin made lethal to tumor cells.
REFERENCES
- 1. Larsson S. C., Kumlin M., Ingelman-Sundberg M., Wolk A. (2004) Dietary long-chain n-3 fatty acids for the prevention of cancer: a review of potential mechanisms. Am. J. Clin. Nutr. 79, 935–945 [DOI] [PubMed] [Google Scholar]
- 2. Amri E. Z., Bertrand B., Ailhaud G., Grimaldi P. (1991) Regulation of adipose cell differentiation. I. Fatty acids are inducers of the aP2 gene expression. J. Lipid Res. 32, 1449–1456 [PubMed] [Google Scholar]
- 3. Anderson M. P., Welsh M. J. (1990) Fatty acids inhibit apical membrane chloride channels in airway epithelia. Proc. Natl. Acad. Sci. U.S.A. 87, 7334–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grimaldi P. A., Knobel S. M., Whitesell R. R., Abumrad N. A. (1992) Induction of aP2 gene expression by nonmetabolized long-chain fatty acids. Proc. Natl. Acad. Sci. U.S.A. 89, 10930–10934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Distel R. J., Robinson G. S., Spiegelman B. M. (1992) Fatty acid regulation of gene expression. Transcriptional and post-transcriptional mechanisms. J. Biol. Chem. 267, 5937–5941 [PubMed] [Google Scholar]
- 6. Bhatt A. P., Jacobs S. R., Freemerman A. J., Makowski L., Rathmell J. C., Dittmer D. P., Damania B. (2012) Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc. Natl. Acad. Sci. U.S.A. 109, 11818–11823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hotamisligil G. S. (2006) Inflammation and metabolic disorders. Nature 444, 860–867 [DOI] [PubMed] [Google Scholar]
- 8. Das U. N. (2005) Long-chain polyunsaturated fatty acids, endothelial lipase, and atherosclerosis. Prostaglandins Leukot. Essent. Fatty Acids 72, 173–179 [DOI] [PubMed] [Google Scholar]
- 9. Calder P. C. (1997) n-3 polyunsaturated fatty acids and cytokine production in health and disease. Ann. Nutr. Metab. 41, 203–234 [DOI] [PubMed] [Google Scholar]
- 10. Pohl J., Ring A., Ehehalt R., Schulze-Bergkamen H., Schad A., Verkade P., Stremmel W. (2004) Long-chain fatty acid uptake into adipocytes depends on lipid raft function. Biochemistry 43, 4179–4187 [DOI] [PubMed] [Google Scholar]
- 11. Covey S. D., Brunet R. H., Gandhi S. G., McFarlane N., Boreham D. R., Gerber G. E., Trigatti B. L. (2007) Cholesterol depletion inhibits fatty acid uptake without affecting CD36 or caveolin-1 distribution in adipocytes. Biochem. Biophys. Res. Commun. 355, 67–71 [DOI] [PubMed] [Google Scholar]
- 12. Hajri T., Abumrad N. A. (2002) Fatty acid transport across membranes. Relevance to nutrition and metabolic pathology. Annu. Rev. Nutr. 22, 383–415 [DOI] [PubMed] [Google Scholar]
- 13. Håkansson A., Zhivotovsky B., Orrenius S., Sabharwal H., Svanborg C. (1995) Apoptosis induced by a human milk protein. Proc. Natl. Acad. Sci. U.S.A. 92, 8064–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Svensson M., Håkansson A., Mossberg A. K., Linse S., Svanborg C. (2000) Conversion of α-lactalbumin to a protein inducing apoptosis. Proc. Natl. Acad. Sci. U.S.A. 97, 4221–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Svanborg C., Agerstam H., Aronson A., Bjerkvig R., Düringer C., Fischer W., Gustafsson L., Hallgren O., Leijonhuvud I., Linse S., Mossberg A.-K., Nilsson H., Pettersson J., Svensson M. (2003) HAMLET kills tumor cells by an apoptosis-like mechanism. Cellular, molecular, and therapeutic aspects. Adv. Cancer Res. 88, 1–29 [DOI] [PubMed] [Google Scholar]
- 16. Fischer W., Gustafsson L., Mossberg A.-K., Gronli J., Mork S., Bjerkvig R., Svanborg C. (2004) Human α-lactalbumin made lethal to tumor cells (HAMLET) kills human glioblastoma cells in brain xenografts by an apoptosis-like mechanism and prolongs survival. Cancer Res. 64, 2105–2112 [DOI] [PubMed] [Google Scholar]
- 17. Gustafsson L., Leijonhufvud I., Aronsson A., Mossberg A.-K., Svanborg C. (2004) Treatment of Skin Papillomas with Topical α-Lactalbumin-Oleic Acid. N. Engl. J. Med. 350, 2663–2672 [DOI] [PubMed] [Google Scholar]
- 18. Mossberg A.-K., Hou Y., Svensson M., Holmqvist B., Svanborg C. (2010) HAMLET treatment delays bladder cancer development. J. Urol. 183, 1590–1597 [DOI] [PubMed] [Google Scholar]
- 19. Mossberg A.-K., Wullt B., Gustafsson L., Månsson W., Ljunggren E., Svanborg C. (2007) Bladder cancers respond to intravesical instillation of HAMLET (human α-lactalbumin made lethal to tumor cells). Int. J. Cancer 121, 1352–1359 [DOI] [PubMed] [Google Scholar]
- 20. Puthia M., Storm P., Nadeem A., Hsiung S., Svanborg C. (2013) Prevention and treatment of colon cancer by peroral administration of HAMLET (human α-lactalbumin made lethal to tumour cells). Gut 0, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aits S., Gustafsson L., Hallgren O., Brest P., Gustafsson M., Trulsson M., Mossberg A.-K., Simon H.-U., Mograbi B., Svanborg C. (2009) HAMLET (human α-lactalbumin made lethal to tumor cells) triggers autophagic tumor cell death. Int. J. Cancer 124, 1008–1019 [DOI] [PubMed] [Google Scholar]
- 22. Storm P., Aits S., Puthia M. K., Urbano A., Northen T., Powers S., Bowen B., Chao Y., Reindl W., Lee D. Y., Sullivan N. L., Zhang J., Trulsson M., Yang H., Watson J. D., Svanborg C. (2011) Conserved features of cancer cells define their sensitivity to HAMLET-induced death. c-Myc and glycolysis. Oncogene 30, 4765–4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Storm P., Klausen T. K., Trulsson M., Ho CS J., Dosnon M., Westergren T., Chao Y., Rydström A., Yang H., Pedersen S. F., Svanborg C. (2013) A unifying mechanism for tumor cell death by ion channel activation. Plos ONE 8, e58578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gustafsson L., Aits S., Onnerfjord P., Trulsson M., Storm P., Svanborg C. (2009) Changes in proteasome structure and function caused by HAMLET in tumor cells. PLoS One 4, e5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Düringer C., Hamiche A., Gustafsson L., Kimura H., Svanborg C. (2003) HAMLET interacts with histones and chromatin in tumor cell nuclei. J. Biol. Chem. 278, 42131–42135 [DOI] [PubMed] [Google Scholar]
- 26. Svensson M., Mossberg A.-K., Pettersson J., Linse S., Svanborg C. (2003) Lipids as cofactors in protein folding. Stereo-specific lipid-protein interactions are required to form HAMLET (human α-lactalbumin made lethal to tumor cells). Protein Sci. 12, 2805–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lisková K., Kelly A. L., O'Brien N., Brodkorb A. (2010) Effect of denaturation of α-lactalbumin on the formation of BAMLET (bovine α-lactalbumin made lethal to tumor cells). J. Agric. Food Chem. 58, 4421–4427 [DOI] [PubMed] [Google Scholar]
- 28. Permyakov S. E., Knyazeva E. L., Khasanova L. M., Fadeev R. S., Zhadan A. P., Roche-Hakansson H., Håkansson A. P., Akatov V. S., Permyakov E. A. (2012) Oleic acid is a key cytotoxic component of HAMLET-like complexes. Biol. Chem. 393, 85–92 [DOI] [PubMed] [Google Scholar]
- 29. Vukojević V., Bowen A. M., Wilhelm K., Ming Y., Ce Z., Schleucher J., Hore P. J., Terenius L., Morozova-Roche L. A. (2010) Lipoprotein complex of equine lysozyme with oleic acid (ELOA) interactions with the plasma membrane of live cells. Langmuir 26, 14782–14787 [DOI] [PubMed] [Google Scholar]
- 30. Tolin S., De Franceschi G., Spolaore B., Frare E., Canton M., Polverino de Laureto P., Fontana A. (2010) The oleic acid complexes of proteolytic fragments of α-lactalbumin display apoptotic activity. FEBS J. 277, 163–173 [DOI] [PubMed] [Google Scholar]
- 31. Cistola D. P., Hamilton J. A., Jackson D., Small D. M. (1988) Ionization and phase behavior of fatty acids in water. Application of the Gibbs phase rule. Biochemistry 27, 1881–1888 [DOI] [PubMed] [Google Scholar]
- 32. Kanicky J. R., Shah D. O. (2002) Effect of degree, type, and position of unsaturation on the pKa of long-chain fatty acids. J. Colloid Interface Sci. 256, 201–207 [DOI] [PubMed] [Google Scholar]
- 33. Lišková K., Auty M. A. E., Chaurin V., Min S., Mok K. H., O'Brien N., Kelly A. L., Brodkorb A. (2011) Cytotoxic complexes of sodium oleate with β-lactoglobulin. Eur. J. Lipid Sci. Technol. 113, 1207–1218 [Google Scholar]
- 34. Louis-Jeune C., Andrade-Navarro M. A., Perez-Iratxeta C. (2011) Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins 80, 374–381 [DOI] [PubMed] [Google Scholar]
- 35. Schnars U., Jüptner W. (2005) Digital Holography: Digital Hologram Recording, Numerical Reconstruction, and Related Techniques, Springer, Berlin [Google Scholar]
- 36. Gustafsson M., Sebesta M., Bengtsson B., Pettersson S. G., Egelberg P., Lenart T. (2004) High-resolution digital transmission microscopy. A Fourier holography approach. Opt. Lasers Eng. 41, 553–563 [Google Scholar]
- 37. Cuche E., Bevilacqua F., Depeursinge C. (1999) Digital holography for quantitative phase-contrast imaging. Opt. Lett. 24, 291–293 [DOI] [PubMed] [Google Scholar]
- 38. Sebesta M., Gustafsson M. (2005) Object characterization with refractometric digital Fourier holography. Opt. Lett. 30, 471–473 [DOI] [PubMed] [Google Scholar]
- 39. Dubois F., Yourassowsky C., Monnom O., Legros J. C., Debeir O., Van Ham P., Kiss R., Decaestecker C. (2006) Digital holographic microscopy for the three-dimensional dynamic analysis of in vitro cancer cell migration. J. Biomed. Opt. 11, 054032 [DOI] [PubMed] [Google Scholar]
- 40. Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID. Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 42. Nozaka M., Kuwajima K., Nitta K., Sugai S. (1978) Detection and characterization of the intermediate on the folding pathway of human α-lactalbumin. Biochemistry 17, 3753–3758 [DOI] [PubMed] [Google Scholar]
- 43. Permyakov E. A., Morozova L. A., Burstein E. A. (1985) Cation binding effects on the pH, thermal, and urea denaturation transitions in α-lactalbumin. Biophys. Chem. 21, 21–31 [DOI] [PubMed] [Google Scholar]
- 44. Brest P., Gustafsson M., Mossberg A.-K., Gustafsson L., Duringer C., Hamiche A., Svanborg C. (2007) Histone deacetylase inhibitors promote the tumoricidal effect of HAMLET. Cancer Research 67, 11327–11334 [DOI] [PubMed] [Google Scholar]
- 45. Pettersson J., Mossberg A. K., Svanborg C. (2006) α-Lactalbumin species variation, HAMLET formation, and tumor cell death. Biochem. Biophys. Res. Commun. 345, 260–270 [DOI] [PubMed] [Google Scholar]
- 46. Pettersson-Kastberg J., Aits S., Gustafsson L., Mossberg A., Storm P., Trulsson M., Persson F., Mok K. H., Svanborg C. (2009) Can misfolded proteins be beneficial? The HAMLET case. Ann. Med. 41, 162–176 [DOI] [PubMed] [Google Scholar]
- 47. Wilhelm K., Darinskas A., Noppe W., Duchardt E., Mok K. H., Vukojević V., Schleucher J., Morozova-Roche L. A. (2009) Protein oligomerization induced by oleic acid at the solid-liquid interface. Equine lysozyme cytotoxic complexes. FEBS J. 276, 3975–3989 [DOI] [PubMed] [Google Scholar]
- 48. Baumann A., Gjerde A. U., Ying M., Svanborg C., Holmsen H., Glomm W. R., Martinez A., Halskau O. (2012) HAMLET forms annular oligomers when deposited with phospholipid monolayers. J. Mol. Biol. 418, 90–102 [DOI] [PubMed] [Google Scholar]
- 49. Mossberg A.-K., Puchades M., Halskau Ø., Baumann A., Lanekoff I., Chao Y., Martinez A., Svanborg C., Karlsson R. (2010) HAMLET interacts with lipid membranes and perturbs their structure and integrity. PLoS One 5, e9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zherelova O. M., Kataev A. A., Grishchenko V. M., Knyazeva E. L., Permyakov S. E., Permyakov E. A. (2009) Interaction of antitumor α-lactalbumin-oleic acid complexes with artificial and natural membranes. J. Bioenerg. Biomembr. 41, 229–237 [DOI] [PubMed] [Google Scholar]
- 51. Hsu P. P., Sabatini D. M. (2008) Cancer cell metabolism. Warburg and beyond. Cell 134, 703–707 [DOI] [PubMed] [Google Scholar]
- 52. Menendez J. A., Lupu R. (2007) Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–777 [DOI] [PubMed] [Google Scholar]
- 53. Nomura D. K., Long J. Z., Niessen S., Hoover H. S., Ng S. W., Cravatt B. F. (2010) Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mashek D. G., Coleman R. A. (2006) Cellular fatty acid uptake. The contribution of metabolism. Curr. Opin. Lipidol. 17, 274–278 [DOI] [PubMed] [Google Scholar]
- 55. Le A., Lane A. N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C. J., Slusher B. S., Zhang H., Zimmerman L. J., Liebler D. C., Slebos R. J., Lorkiewicz P. K., Higashi R. M., Fan T. W., Dang C. V. (2012) Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 15, 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang W. C., Shyh-Chang N., Yang H., Rai A., Umashankar S., Ma S., Soh B. S., Sun L. L., Tai B. C., Nga M. E., Bhakoo K. K., Jayapal S. R., Nichane M., Yu Q., Ahmed D. A., Tan C., Sing W. P., Tam J., Thirugananam A., Noghabi M. S., Pang Y. H., Ang H. S., Mitchell W., Robson P., Kaldis P., Soo R. A., Swarup S., Lim E. H., Lim B. (2012) Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148, 259–272 [DOI] [PubMed] [Google Scholar]
- 57. Svensson M., Fast J., Mossberg A.-K., Düringer C., Gustafsson L., Hallgren O., Brooks C. L., Berliner L., Linse S., Svanborg C. (2003) α-Lactalbumin unfolding is not sufficient to cause apoptosis, but is required for the conversion to HAMLET (human α-lactalbumin made lethal to tumor cells). Protein Sci. 12, 2794–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pettersson-Kastberg J., Mossberg A. K., Trulsson M., Yong Y. J., Min S., Lim Y., O'Brien J. E., Svanborg C., Mok K. H. (2009) α-Lactalbumin, engineered to be nonnative and inactive, kills tumor cells when in complex with oleic acid. A new biological function resulting from partial unfolding. J. Mol. Biol. 394, 994–1010 [DOI] [PubMed] [Google Scholar]