Background: Oligopeptidases are serine proteases cleaving only short peptides.
Results: The complex channel system found within a hexameric oligopeptidase presents a rigid, double-gated model for size-based substrate selection.
Conclusion: The substrate selection mechanism applied by an oligopeptidase depends on its multimerization state.
Significance: Degradation of cytotoxic and misfolded proteins is aided by oligopeptidases, which are thus possible targets of cancer therapy.
Keywords: Aggregation, Peptide Interactions, Protein Self-assembly, Serine Protease, X-ray Crystallography, Acylaminoacyl Peptidase, Amylogenic β-Strand, Self-compartmentalization, Substrate Size Selection
Abstract
Oligopeptidases impose a size limitation on their substrates, the mechanism of which has long been under debate. Here we present the structure of a hexameric serine protease, an oligopeptidase from Pyrococcus horikoshii (PhAAP), revealing a complex, self-compartmentalized inner space, where substrates may access the monomer active sites passing through a double-gated “check-in” system, first passing through a pore on the hexamer surface and then turning to enter through an even smaller opening at the monomers' domain interface. This substrate screening strategy is unique within the family. We found that among oligopeptidases, a residue of the catalytic apparatus is positioned near an amylogenic β-edge, which needs to be protected to prevent aggregation, and we found that different oligopeptidases use different strategies to achieve such an end. We propose that self-assembly within the family results in characteristically different substrate selection mechanisms coupled to different multimerization states.
Introduction
Members of the prolyl oligopeptidase family are serine proteases, such as prolyl oligopeptidase (POP)5 itself, dipeptidyl peptidase IV (DPP-IV), oligopeptidase B (OPB), and acylaminoacyl peptidase (AAP), that are able to distinguish between potential substrates based on their size and only cleave those that do not exceed 30 amino acids in length (1, 2). When the first crystal structure of a serine oligopeptidase, that of POP, was determined (3), size exclusion was explained by the domain structure of the enzyme, which later proved to be quite similar for all family members. Oligopeptidases consist of a half-sphere-shaped hydrolase region containing the active site and a cylindrical propeller domain, which caps it, burying the Ser-His-Asp catalytic triad in a well protected inner hole with dimensions comparable with the size limitations imposed by the enzyme. The structure, however, raised the question of substrate access and, through it, how enzyme effectiveness and selectivity are simultaneously maintained in the case of such a decidedly buried active site.
It was initially proposed that substrates must reach the active site through the narrow channel bisecting the propeller domain. Alternatively, substrate access routes must target the domain interface, either through a transient opening between the propeller and the hydrolase domains or through a permanent hole between the two.
POP, the first discovered and most extensively studied member of the family, was revealed to have both a closed (4)6 (PDB codes 1H2W and 3MUN) and an open arrangement (6, 7) (PDB codes 3IUL and 1YR2) in crystal structures of its unligated form, in the latter of which the cylindrical dome of the propeller region flips back, exposing the active site entirely. This suggests that an open/closed dynamic equilibrium, fine-tuned by the amino acid composition of the interacting domain surfaces, might describe the resting state of the enzyme, where both forms coexist. This notion recently gained support by the determination of the crystal structure of another oligopeptidase, AAP from Aeropyrum pernix (ApAAP), which contains both the open and closed forms of the unligated enzyme within the same crystal lattice, in the form of 1:1 open/closed mixed dimers (8). In both cases, a keen control mechanism is coupled to the opening; the active site is disassembled in the open form mainly by the destabilization and reformation of the flexible loop containing the catalytic His (6, 8). Substrates exceeding the size limit disrupt the closing of the enzyme and thus only encounter the impaired active site of the open form and leave the transient complex unchanged.
An alternate possible substrate selection strategy is formation of a permanent entrance hole and providing for its shielding. DPP-IV was shown to have such an architecture; substrates can access its active site either through its large propeller hole (9) or through a spacious side opening between its hydrolase and propeller domains stabilized and partly covered by dimerization (10, 11, 12).
In this work, we present an even more complex example of the latter: that of acylaminoacyl peptidase from Pyrococcus horikoshii (PhAAP), which we found active in the hexameric form (13). Three crystal structures were determined, which demonstrates that hexamerization results in self-compartmentalizing, a self-assembled double-gated channel system that may effectively screen the substrates of the six individual monomers, all possessing a permanent entry at the domain interface. Although hexameric arrangement is unique among classical serine proteases, it is reminiscent of other self-compartmentalizing intracellular proteases, such as Gal6/bleomycin hydrolase (14), tricorn protease (15), Lon protease (16), or the complex systems of proteasomes.
Thus, the PhAAP structure provides a unique possibility of comparison among oligopeptidases sharing common domain structure but applying different substrate selection strategy and among self-compartmentalizing proteases of different monomeric structure but of similar selection strategy. This comparison allowed us to propose that the flexibility of the loop holding the catalytic His residue, the accessibility of a sticky β-strand anchoring the His-loop, and unobtrusive insertions or terminal extensions determine the substrate selection strategy and self-assembly in the oligopeptidase family. Structure determination of PhAAP and understanding of the structural requirements and consequences of multimerization also takes us one step closer to the physiologically relevant mammalian enzyme, which is active as a tetramer.
AAP functions as an exopeptidase in mammals, removing N-terminally blocked amino acids from peptides, although other orthologs have endopeptidase activity too (13, 17). It has been shown to act in concert with and exert regulation over the proteasome (18) while also being implicated in renal and small cell carcinoma (19–21), and it is also referred to as a potential target of certain cognitive enhancers (22, 23).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification for Crystallization
PhAAP was expressed in Escherichia coli and purified as described previously for PhAAP and ApAAP (13, 17). The active site mutation was introduced in PhAAP (S466A) by the two-step PCR method as described for ApAAP (17). The protein solutions were concentrated to 10 mg/ml in 20 mm Tris/HCl buffer (pH 8.0). Preparing the covalent complex of PhAAP with the inhibitor benzyloxycarbonyl-glycyl-glycyl-phenylalanyl-chloromethyl ketone (CMK) was carried out by adding the inhibitor in 2.4-fold excess to the protein solution.
Crystallization, Data Collection, Phasing, and Refinement
The PhAAP structure was determined from crystals grown at 20 °C by the hanging drop method. Crystal quality was improved by microseeding. The crystals were flash-frozen and stored in liquid nitrogen prior to data collection.
For the native crystal, drops were prepared by mixing 2 μl of protein solution and 2 μl of reservoir solution (3.0 m 1,6-hexanediol, 0.20 m MgCl2, 0.1 m Tris/HCl buffer, pH 8.5). The protein solutions contained 10.5–13.0 mg/ml PhAAP in 20 mm Tris/HCl buffer (pH 8.0). Phase information was obtained using the following three derivatives. A crystal was soaked in 4.5 μl of reservoir solution (same as for the native crystal) mixed with 1.5 μl of 0.05 m UO2(NO3)2 solution for 1 h prior to flash cooling. A crystal was soaked in 2 μl of reservoir solution mixed with 2 μl of 0.02 m K2PtCl4 solution for 2 h prior to flash cooling. After mixing 2 μl of 0.2 m KI solution in the crystallization drop, a crystal was quick soaked in it.
The orthorhombic crystal form was obtained by a co-crystallizing trial of the S466A mutant form of PhAAP and its substrate (13) N-benzyloxycarbonyl-Glu-Phe-Ser-Pro-(para-nitro-Phe)-Arg-Ala. The substrate was dissolved in the protein solutions (9.2 mg/ml protein in 20 mm Tris/HCl buffer, pH 8.0) at a 10-fold excess. The drops contained 2 μl of protein solution and 2 μl of reservoir solution (3.0 m 1,6-hexanediol, 0.30 m MgCl2, 0.1 m Tris/HCl buffer, pH 8.0). The structure, however, does not contain any fragment of the substrate, possibly because 1,6-hexanediol competes for and occupies the S1 site. For studying binding of a substrate fragment, we prepared crystals with a covalent inhibitor, CMK.
The crystal of the PhAAP-CMK complex was grown in a hanging drop of 4 μl of the PhAAP-CMK solution (9.7 mg/ml covalent protein-inhibitor complex in 20 mm Tris/HCl buffer, pH 8.0) and 4 μl of reservoir solution (2.5 m 1,6-hexanediol, 0.20 m MgCl2, 0.1 m Tris-HCl buffer, pH 8.5).
Data collection was carried out on a Rigaku diffractometer (Cu Kα radiation, wavelength 1.5418 Å) for the native, UO2(NO3)2, and KI derivative crystals (graphite monochromator, R-AXIS IIc detector) as well as the CMK complex (blue optics, R-AXIS IV++ detector). Data sets of the K2PtCl4 derivative crystal were collected at synchrotron sources at wavelengths 0.8162 and 1.0715 Å, at the EMBL beamline X11 of the Deutsches Elektronen-Synchrotron (DESY) and at beamline BM14 of the European Synchrotron Radiation Facility (ESRF), respectively. The data set of a crystal of the orthorhombic crystal form was collected at beamline BM14 of ESRF (wavelength 1.0715 Å). All data were collected at 100 K. Data processing was carried out using the XDS and XSCALE programs (24) for all data sets. All but one crystal were isostructural with the hexamers of AAP formed by space group symmetry H32 from the single monomer of the asymmetric unit. The asymmetric unit of the crystal in the orthorhombic crystal form contains two hexamers of PhAAP (for this data set, reflections for Rfree calculation were selected in thin shells).
Data sets of the native, UO2(NO3)2 and KI and K2PtCl4 derivatives were used for solving the phase problem using multiple isomorphous replacement with anomalous scattering. The initial set of sites was located by the SHELX package (25). Phases were refined with the program MLPHARE (26), and they were further improved with the program DM (27). An initial model was automatically generated by Buccaneer (28). The phase problem for the orthorhombic crystal form was solved by molecular replacement using the program MOLREP (29). Manual model building was performed for all of the three structures using the program Coot (30). Refinement was carried out either by using REFMAC (31) (native and CMK complex) or Phenix (32) (orthorhombic crystal form), including TLS B-factor refinement for each individual protein domain. The model was validated using SFCHECK (33) and MolProbity (34). The percentages of residues in the favored/disallowed Ramachandran regions are as follows: PhAAP native structure, 95.82%/0.17%; PhAAP-CMK complex, 96.22%/0.33%; PhAAP orthorhombic crystal form, 96.19%/0.00%. The final statistics can be found in Table 1.
TABLE 1.
Data collection, phasing, and refinement statistics (multiple isomorphous replacement with anomalous scattering)
| Native (unligated) (in house) | Uranyl derivative (in house) | Iodine derivative (in house) | Platinum derivative |
CMK inhibitor complex (in house) | Orthorhombic unligated (ESRF BM14) | ||
|---|---|---|---|---|---|---|---|
| DESY X11 | ESRF BM14 | ||||||
| Data collectiona | |||||||
| Space group | H 3 2 | H 3 2 | H 3 2 | H 3 2 | H 3 2 | H 3 2 | P 21 21 21 |
| Cell dimensions | |||||||
| a, b, c (Å) | 183.000, 183.000, 144.632 | 182.953, 182.953, 143.936 | 182.819, 182.819, 144.443 | 184.802, 184.802, 145.727 | 184.802, 184.802, 145.727 | 184.057, 184.057, 145.529 | 183.31, 183.80, 275.74 |
| α, β, γ (degrees) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Wavelength | 1.5418 | 1.5418 | 1.5418 | 0.8162 | 1.0715 | 1.5418 | 1.0715 |
| Resolution (Å)b | 20.0-1.90 (2.00-1.90) | 20.0-2.5 (3.0-2.5) | 20.0-2.25 (2.5-2.25) | 20.0-2.00 (2.15-2.00) | 20.0-2.50 (2.80-2.50) | 20.0-1.60 (1.64-1.60) | 20.0-2.65 (2.72-2.65) |
| Rmeasb | 0.158 (0.686) | 0.141 (0.319) | 0.138 (0.357) | 0.112 (0.414) | 0.071 (0.212) | 0.066 (0.738) | 0.090 (0.690) |
| I/σIb | 12.79 (1.87) | 10.94 (5.18) | 10.22 (3.74) | 15.35 (5.02) | 30.97 (12.35) | 22.25 (2.57) | 16.47 (2.36) |
| Completeness (%)b | 95.7 (74.6) | 99.2 (100.0) | 98.8 (99.1) | 99.4 (97.4) | 99.5 (99.0) | 99.2 (94.4) | 94.9 (96.8) |
| Redundancyb | 8.32 (2.84) | 4.05 (4.07) | 2.80 (2.66) | 6.10 (5.96) | 11.53 (11.43) | 7.03 (5.24) | 4.39 (4.33) |
| Refinement | |||||||
| Resolution (Å) | 19.78-1.91 | 19.74-1.60 | 19.98-2.70 | ||||
| No. reflectionsc | 64,801 (3425) | 116,219 (6218) | 222,127 (1751) | ||||
| Rwork/Rfree | 0.172/0.210 | 0.155/0.186 | 0.200/0.241 | ||||
| No. of monomers/asymmetric unit | 1 | 1 | 12 | ||||
| No. atoms | |||||||
| Protein | 5082 | 5093 | 58,518 | ||||
| Ligand/ion | 51 | 101 | 231 | ||||
| Water | 395 | 697 | 1161 | ||||
| B-Factors | |||||||
| Protein | 21.23 | 21.33 | 67.98 | ||||
| Ligand/ion | 39.16 | 40.05 | 57.74 | ||||
| Water | 28.73 | 33.89 | 45.22 | ||||
| Root mean square deviations | |||||||
| Bond lengths (Å) | 0.018 | 0.025 | 0.005 | ||||
| Bond angles (degrees) | 1.944 | 2.248 | 0.979 | ||||
a Each data set was collected from one crystal.
b Values in parentheses are for the highest resolution shell.
c The reflection number used in the Rfree calculation is shown in parenthesis.
Molecular Dynamics (MD) Simulations
60-ns NPT MD simulations were carried out on monomers of POP (using PDB structure 1H2W (4) as a starting conformation) and PhAAP (using the presently determined native structure as starting conformation) at 300 K. The systems were pre-equilibrated at 300 K by decreasing the restraints on the protein atoms and submitting the free system to a 200-ps NVT step to stabilize pressure. The GROMACS program suite (35) and CHARMM27 force field (36, 37) were utilized. Systems were solvated by ∼28,000 TIP3P waters, protein overall charge was neutralized, and physiological salt concentration was set using Na+ and Cl− ions. Average geometries and B-factors were calculated using the last 10 ns of the trajectories. Figures were created using PyMOL (v0.99, DeLano Scientific, 2006) and Maestro version 9.2 (Schrodinger, LLC, New York).
RESULTS
Crystal Structures
Crystal structure of the PhAAP hexamer was determined in two crystal forms. Two sets of data originate from crystals bearing R32 space group symmetry, where the six monomers of the hexamer are crystallographically equivalent. One of these shows the substrate-free form of the enzyme, whereas the other contains a chloromethyl ketone inhibitor (CMK) covalently bound to the active site serine and histidine residues, serving as an analog of the intermediate of the substrate hydrolysis reaction. The second crystal form is orthorhombic, containing two substrate-free hexamers in the asymmetric unit, with no crystallographic restrictions for the symmetry and topology of the monomers within the hexamer (Table 1). The monomer units within each hexamer and in between monomers of the different structures are strikingly similar (after fitting to the native unligated structure, root mean square deviation of Cα atoms is 0.23 Å for the CMK complex structure, and it is between 0.25 and 0.41 Å for the 12 molecules of the orthorhombic structure, respectively). Furthermore, the overall hexameric form is also quite similar in all of the three structures, indicating that the hexamer is a symmetric object with a well defined structure.
Overall Structure of the PhAAP Monomer
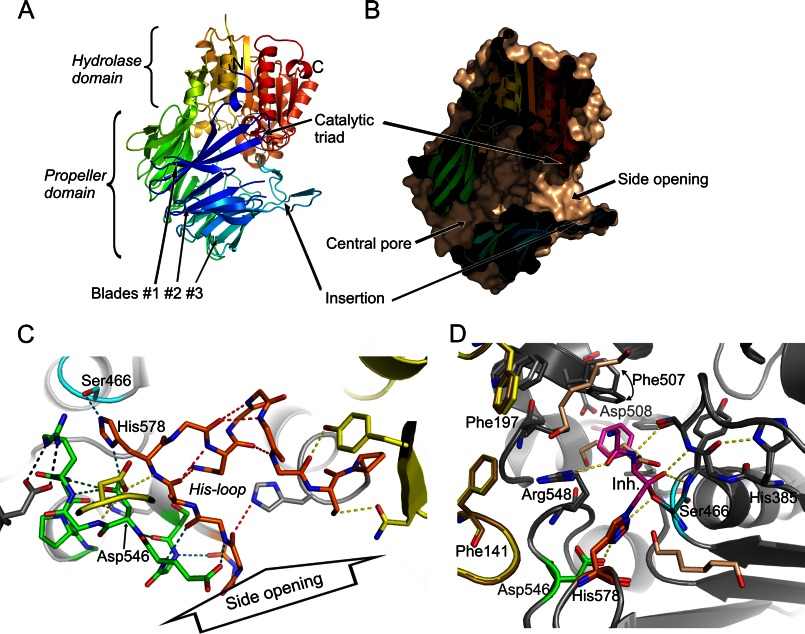
The enzyme monomer consists of a dome-shaped, seven-bladed β-propeller domain and the C-terminal catalytic domain of α/β-hydrolase fold. Each blade of the β-propeller domain contains four antiparallel β-strands. There is a large cavity with overall diameter of about 22 Å between the two domains, which contains the active site (Fig. 1, A and B).
FIGURE 1.
Structure of PhAAP reveals two entrances to the interdomain cavity. The structure of the monomer is shown in a schematic representation (A), and its cross-section is shown with a molecular surface (wheat) (B). The two entrances to the central cavity of the monomer are shown (the pore of the β-propeller domain and the side opening formed between blade 1 and insertion of blade 3 of the propeller domain). C, the loops holding the catalytic triad (Ser466, His578, and Asp546; light blue, orange, and green, respectively) are exposed to solvent and form only few contacts with the propeller domain (yellow), yet they are stabilized by intra- and interloop hydrogen bonds. (Hydrogen bonds are color coded as follows. Red, intra-His-loop; green, intra-Asp-loop; light blue, between His- and Asp-loops; black, formed with the rest of the hydrolase domain; yellow, with the propeller domain.) D, comparison of the PhAAP structure with hexanediol (wheat) bound in the substrate specificity pocket and that of PhAAP in complex with a covalently bound chloromethyl ketone inhibitor (magenta; with two hexanediol molecules outside the S1-S2 region shown in wheat) reveals that the only significant difference is adjustment of the S1 pocket by rotation of the Phe507 side chain in the covalent complex (in darker colors).
A side opening of this cavity (12 × 16 Å) is created under blade 2, gated by blades 1 and 3 (Fig. 1B). Blade 2 is both shortened and shifted upward by 6–8 Å as compared with the corresponding blades of other family members, such as POP, OPB, or ApAAP, but similar to that of DPP-IV with an even greater interdomain opening of 15 × 22 Å formed by shortening and shift of its blades 1–3 (9).
The single structure homologous to PhAAP available to date is that of the β-propeller domain of trilobed protease from Pyrococcus furiosus (TLP, sequence identity 79 and 85% for the propeller domain and the full protein chain, respectively (PDB code 2GOP (38)). Alignment of the propeller domains of PhAAP and TLP reveals high structural similarity (root mean square deviation of fitted Cα atoms is 0.94–1.06 Å), and the conformational differences as well as disorder of certain loop regions of TLP can all be explained by the fact that in PhAAP, these segments are involved in interactions with the hydrolase domain (which is missing from the TLP structure) or other monomers of the PhAAP multimer structure (supplemental Fig. 1).
The hydrolase domain (residues 1–14 and 347–618) that contains the catalytic triad, consists of a short N-terminal segment and the residues sequentially following the seven-bladed β-propeller domain inserted between the two (residues 15–346). The N-terminal segment is unusually short within the family; it contains only 14 amino acid residues (as compared with the 22, 31, 72, and 94 amino acids of the same region of ApAAP, DPP-IV, POP, and OPB, respectively), whereas the structure of the core hydrolase domain is well conserved despite its relatively low sequence homology.
In PhAAP, the Ser466 of the catalytic triad is positioned on a short turn between a β-strand, member of an eight-stranded β-sheet, and a helix and is thus in a rather well defined environment. This feature is conserved among oligopeptidases, as is the fact that the other two members of the catalytic triad, His578 and Asp546, are placed on neighboring, longer loops (His-loop/Asp-loop), connecting β-stands of the very same β-sheet that the Ser is linked to, with helices that stand on either side of the sheet, at the point where it reaches the surface of the hydrolase domain (Fig. 1C). Thus, the catalytic His and Asp residues provide direct coupling between the rather buried catalytic Ser and the domain surface. The Asp-loop (residues 543–552) is well structured with four intraloop hydrogen bonds, similarly to the corresponding loops of the other family members. However, the well defined nature of the His-loop of PhAAP (residues 574–586) is unique; there are six intraloop hydrogen-bonds contributing to its internal stability (Fig. 1C). The only other example of a rigidified His-loop among oligopeptidases is that of DPP-IV, with four hydrogen bonds in this region. In the rest, 0–2 intraloop hydrogen bonds can be found in the corresponding His-loops (zero in ApAAP (8), one in POP from porcine brain (4) and muscle (3) and from Aeromonas caviae,6 two in POP from Myxococcus xanthus (39) and OPB (40); PDB codes are 3o4j, 1h2w, 1qfm, 3mun, 2bkl, and 2xe4, respectively); their conformations are mainly defined by their interaction with the neighboring Asp-loops and by interdomain connections with the loops of the propeller domain. In PhAAP, five hydrogen bonds link the His-loop and the Asp-loop to one another in addition to the catalytic His578-Asp546 interaction, but neither the His- nor the Asp-loop forms direct interdomain hydrogen bonds with the propeller domain. This is the case because, interestingly, these two loops line the entrance hole facing (through the cavity) the shortened blade 2 of the propeller, so besides participating in the catalytic reaction, these regions may partake in the screening and direction of the substrates too.
Accommodation of the Covalently Bound Inhibitor and Hexanediol Molecules in the Active Site
In both crystal forms of the substrate-free enzyme, the substrate specificity pocket was occupied by 1,6-hexanediol (Fig. 1D), which was present in the crystallization condition in high concentration (3 mol/dm3). For studying substrate binding, a covalent adduct of PhAAP and a chloromethyl ketone inhibitor was crystallized, and its structure was solved. The covalently bound part and the Phe and Gly moieties occupying the S1 and S2 subsites are well defined by electron density and are bound in canonical position (supplemental Fig. 2). The N-terminal benzyloxycarbonyl-glycyl moiety, however, cannot be located in the electron density map, suggesting that it does not specifically bind to the enzyme surface. Comparison of the structure containing the covalently bound inhibitor mimicking the reaction intermediate and that of the substrate-free form reveals high similarity of the two, indicating that the substrate binding site and the catalytic apparatus are preformed in the latter.
The Active Site Structure Is Consistent with Endopeptidase Activity
The active site structure of PhAAP resembles that of ApAAP (Fig. 1D). The catalytic triad and the oxyanion site display an active conformation in both enzymes. The conformation and interactions of the loop holding the conserved His stabilizing the oxyanion hole (His367) highly resemble that of ApAAP, because it was proposed that this structural element is characteristic of all AAPs within the prolyl oligopeptidase family (41). The substrate specificity pocket (S1) is deep, like in ApAAP (supplemental Fig. 3), allowing the binding of large hydrophobic side chains, such as Phe or Leu. Most of the residues accommodating the P1 side chain are hydrophobic, and most of them are conserved or similar in PhAAP and ApAAP (Trp494/Trp474, Val491/Val471, Tyr467/Tyr446, Val511/Leu492, Phe507/Phe488, Ser501/Ser481, and Ile503/Phe485). However, an interesting difference can be observed. In the bottom of the S1 pocket of PhAAP, a small polar (Ser497) and a negatively charged residue (Asp508) are located, whereas in ApAAP, the corresponding side chains are Met477 and Ile489, respectively, suggesting that PhAAP may also cleave after positively charged residues (supplemental Fig. 3). Indeed, we observed that in the absence of Phe at the P1 site, substrates with Arg or Lys can also be hydrolyzed (data not shown).
In addition to the acylaminoacyl peptidase activity, PhAAP also exhibits endopeptidase activity (13). The structure is in line with this finding. The substrate binding site is extended and does not have any steric or electrostatic blocking at the S2-S3 sites, which true exopeptidases, like DPP-IV, do.
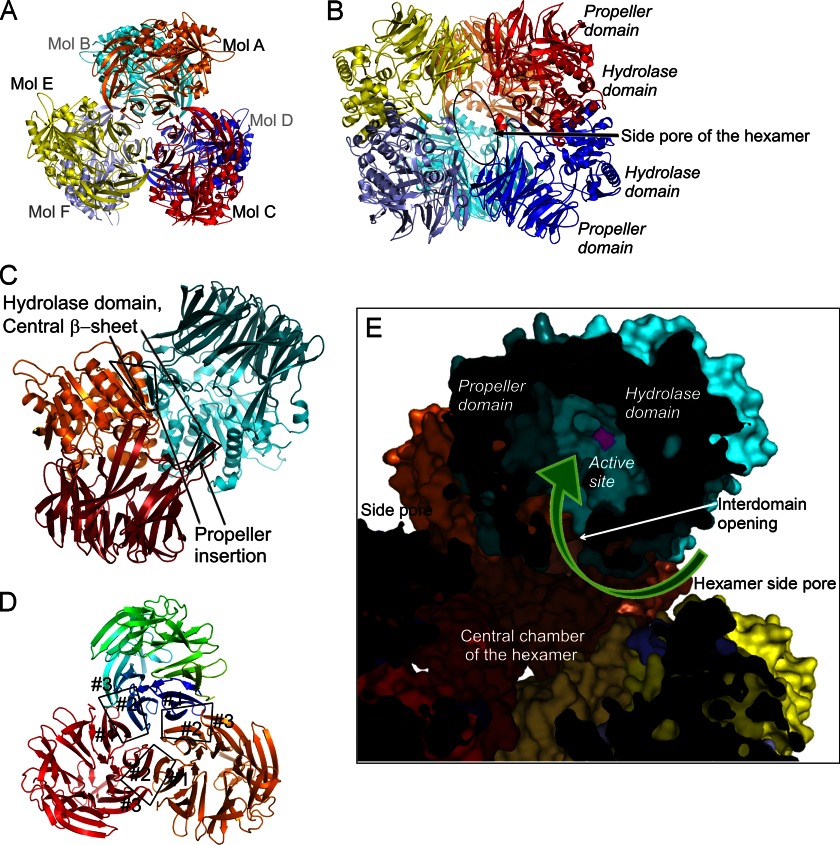
Stabilization of the PhAAP Hexameric Structure; Trimer of Dimers
In a previous study, we have shown, using size exclusion chromatography, that PhAAP is a homohexameric enzyme in solution (13). Hexamers of PhAAP in the crystalline state are formed as trimers of dimers of the monomer enzyme (Fig. 2, A and B). Analysis of molecular contacts revealed that the association within the dimers is much stronger than that securing trimerization. Both the area of the buried surfaces and the free enthalpy contribution of the dimers dominate over those of the trimers (supplemental Table 1). Although the highly homologous enzyme, TLP was proposed to be trimeric (38), we suggest that it may form hexamers in solution, because the sequence identity between PhAAP and TLP is even higher at the contact regions of the trimer interfaces formed by the propeller domains (81% compared with 79% for the propeller domain) as well as for the dimer interfaces formed by both domains (96%, compared with 85% for the whole protein) (supplemental Table 2).
FIGURE 2.
Structure of the symmetric hexamer of PhAAP allows for substrate selection by a rigid gate system. A and B, top and side views of the hexamer formed as trimer of dimers. There are three large side pores leading to a central chamber of the complex. C, the dimers are stabilized mainly by contacts of the insertion of the propeller domain and the hydrolase domain of the other monomer, forming an extension of the central β-sheet. D, the trimers are stabilized by interactions of blades 1–3 of the propellers. E, cross-section of the hexamer with molecular surfaces of PhAAP molecules (color code is similar to that in A, with the propeller domains in darker shades). Substrate oligopeptides entering the central chamber through the side pores of the hexamer should be flexible enough to pass through the narrower interdomain opening of the monomers to get to the active site.
The dimer interface lies between the long loop insertion of blade 3 from the propeller domain of one monomer and the hydrolase domain of the other monomer (Fig. 2C). The long insertion of one molecule lines up to and extends the central eight-stranded β-sheet holding the catalytic residues of the other, by forming the ninth strand of it in an antiparallel manner (framed in Fig. 2C; supplemental Fig. 4). In contrast, in ApAAP, which is active in the dimeric form, the dimer interface is formed between the two hydrolase domains so that the central β-sheets of the two hydrolase domains are connected by forming a 16-stranded continuous sheet, whereas the propeller domain of one momomer has no contacts with the either domains of the other (supplemental Fig. 4).
The trimer interfaces are composed of the outer side of blade 1 of the propeller of one monomer and the outward facing part of blades 2 and 3 of its neighboring monomer (Fig. 2D, blades 1–3 are labeled for the three monomers) and are dominated by side chain interactions. Blades 1–3 are those lining the entrance of the monomer; thus, trimerization contributes to fixing of the pore size by restricting the blades' upward movement.
Hexamerization creates further substrate size screeners. Three large openings of about 20 × 30 Å can be found on the hexamer surface (Fig. 2, B and E), which continue in tunnels and are joined in a big cavity in the middle of the hexamer. The interdomain opening of each monomer is also connected to this central chamber of the hexamer, because all six cavities face inward. In principle, the substrate can reach one of the active sites in two ways: 1) first entering the opening of the hexamer and subsequently the side chamber of a monomer or 2) through the central pore of the propeller domain, provided that the pore can be widened (42). However, the trimerization contacts restrict the flexibility of the propeller pore region, which makes this second option unlikely (Fig. 2E). The two successive openings and the complex tunnel system between them seem sufficient for screening the substrates. Those molecules that are small enough to enter the antechamber through the larger hole but are not flexible enough for entering the second may simply pass through the central chamber and leave at the opposite end; however, those that can proceed through the monomer cavity to the active site will be cleaved.
MD Simulations for Determining Monomer Stability
MD simulations were carried out to determine the monomer stability of two members of the prolyl oligopeptidase family, those of POP and PhAAP. Calculations were designed to test the significance of multimerization; POP is active and stable as a monomer, whereas PhAAP hexamerizes. Monomer structures were compared after 60 ns of simulation time. The two molecules behave quite similarly; the conformation seen in the crystal structure of each was preserved in the simulation. This has a special significance in the case of the PhAAP structure because it also demonstrates that the 1,6-hexanediol molecule trapped in the active site in the crystalline state and absent in the simulation does not have a structure-determining role. The overall root mean square deviation of main chain atoms from their crystallographically determined position was found to be 1.34 and 1.44 Å in POP and PhAAP, respectively (supplemental Fig. 5).
A notable difference between the crystal structures and the simulation results can be seen in the conformation of the catalytic triad, both in the closed structure of POP and in PhAAP with its wide side opening. The hydrogen-bonded triad of the crystal structures loosens in the fully solvated monomer. The Ser-Oγ–His-Nϵ2 distance increases from 2.9 and 2.6 Å to 4.7 and 4.6 Å, in the case of POP and PhAAP, respectively, due mainly to the active Ser sampling a different rotamer, relaxing its strained, nearly eclipsed N+1-C-Cα-Cβ torsion of 2–5° seen in the crystal structures to that of a more staggered 35°. Ser and His stay connected through a shared water molecule in 58 and 19% of the structures of the last 10 ns of the trajectory in POP and PhAAP, respectively, and through two or more coordinated water molecules in the rest. The His-Asp hydrogen bond remains intact in all snapshots. Such a “latent” conformation of the Ser-His-Asp catalytic triad was seen in the crystal structure of ligand-free tricorn interacting factor F1 (43), where, in the absence of propeller domain, the hydrolase is only partially shielded from solvent. In crystal structures of other oligopeptidases, in addition to the strained backbone conformation of the catalytic Ser, the above mentioned N+1-C-Cα-Cβ torsion is also in a high energy conformation of −6–7°. It seems that the partially “dried out” condition of crystallization or the binding of a substrate is required to force the Ser into the conformation that guarantees its reactivity.
DISCUSSION
When comparing the structure of PhAAP presented here and those of oligopeptidases determined previously, three fundamentally different substrate admission routes can be detected: 1) allowing access of substrates through the propeller hole of the closed form, 2) dynamic domain flapping between the closed and open conformation, or 3) a channel system created by multimerization.
The first provides an almost too strict screener; the narrow pore of the propeller domain cannot be appropriate for all oligopeptidase substrates. However, DPP-IV, for example, with an eight-bladed propeller domain and a correspondingly wider pore than those of POP, ApAAP, OPB, or PhAAP was proposed to function that way (9), although, because it can also cleave even 80-amino acid-long peptides, it must apply other substrate admission routes too (10).
POP and ApAAP seem to utilize the second strategy. In the closed conformation of these enzymes, the hydrolase and propeller domains sit on top of each other, burying their active site. In their open form, the relative domain position is fixed only on one side by a hinge region, whereas at the opposite side of the molecules, the domains flap away from each other, forming a 40–50° opening, causing an ∼20-Å shift at the point of the greatest opening. This conformation provides easy access to the active site. In all cases where such an open form was observed, the active site was disassembled and inactivated by the opening, chiefly through the destabilization of the loop holding the catalytic His residue. In the closed form, this loop is supported by interdomain hydrogen bonds that are lost when the opening takes place and the domains move away from each other (6, 8).
In the crystal structure of PhAAP described in this paper, the third strategy can be witnessed. Hexamerization creates a compartmentalized inner space, with a complex, double-gated “check-in” system. Substrates first have to pass through the entrance at the hexamer surface (Fig. 2E). Once inside, the active sites can be reached through the spacious side opening of the monomers created by the shortening and upward shift of the second of the seven blades of the propeller domains.
The overall topology enables all oligopeptidases to widen their propeller pore in an induced fit step, to multimerize, or to open, thus to utilize any of the three outlined strategies. The interesting question is which structural features encode selecting one or the other.
A crucial feature to be considered is the flexibility of the loop containing the His residue of the catalytic triad (His-loop). Pliability of this region is essential for those oligopeptidases that open up, because selectivity is assured by “switch-off” of the active site in the open form by destabilization of the His-loop. Accordingly, in the cases of POP, OPB, and ApAAP, the shape of the His-loop and the position and orientation of the catalytic His are defined only by interloop contacts, both with the neighboring Asp-loop containing the third member of the triad and with the facing loops of the propeller domain. Therefore, the shape and stability of the His-loop is coupled to the proximity of the propeller domain. In contrast, in multimer enzymes with permanent entrance between the two domains (PhAAP and DPP-IV), the His-loop is rigidified in the active conformation by a number of intraloop hydrogen bonds.
In all oligopeptidase structures so far determined, the His-loop connects an α-helix and the terminal β-strand of the central β-sheet of the hydrolase domain, both at the surface of the monomer. Interestingly, this edge β-strand of 6–8 residues is of regular extended geometry in all cases, without twists, bulges, or prolines to disrupt its hydrogen-bonding potential; thus, it is an ideal aggregation primer (44). The corresponding sequences were recognized by the WALTZ predictor (45) as having high probability for amyloid formation, PhAAP scoring the highest value, 98.3, whereas the others scored between 79.9 and 89.0.
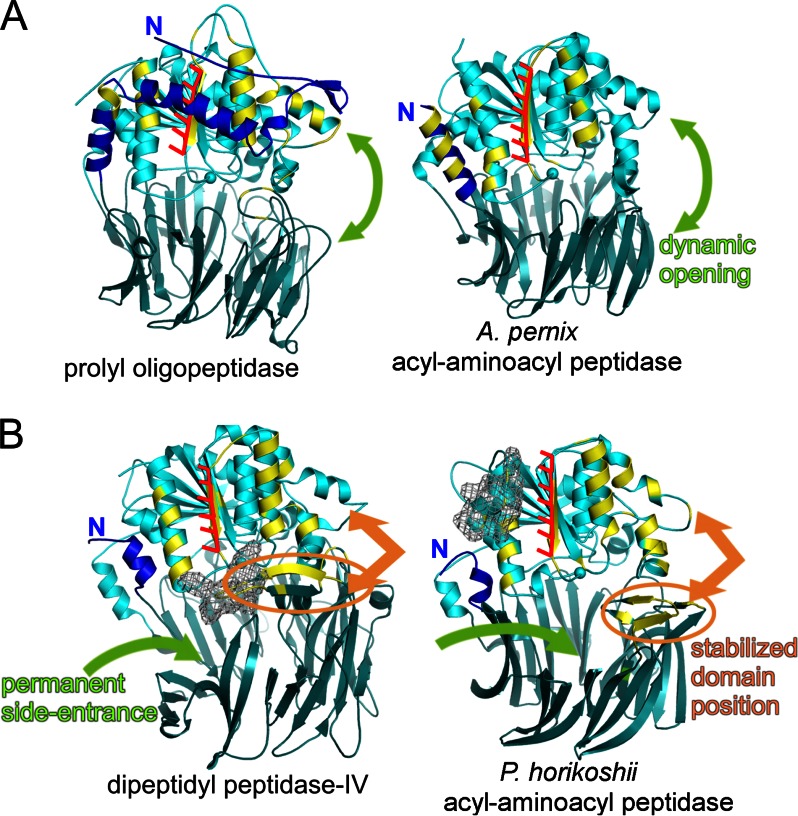
The propensity for amyloid-type aggregation is a generic property of proteins (46), which thus have evolved structural and sequential adaptations to protect their surface-close β-strands (44, 47, 48), because aggregation prompted by such segments, even in the native fold or in locally unfolded states, has been shown to be the first step of toxic aggregation processes (49, 50). Negative selection against conformers with significant tendency to aggregate is now regarded as one of the key determinants in the development of complex molecular machineries (51), as seen in the case of PhAAP in the present study. (Fig. 3).
FIGURE 3.
Strategies of shielding the sticky β-edge of the hydrolase domain to prevent aggregation within the prolyl oligopeptidase family. Hydrolase domain cores are shown in light blue with a red fork at its β-edge. Regions covered by other monomers or segments are colored yellow. The propeller domains are shown in gray-blue. A, in monomeric enzymes, like prolyl oligopeptidase and oligopeptidase-B, a long N-terminal segment (blue) wraps around the hydrolase core and shields the β-edge. In A. pernix AAP dimer, the other monomer covers the β-edge, forming a continuous β-sheet. The propeller domains are not involved in these interactions and remain floppy, allowing dynamic open/closed equilibrium for substrate screening. B, in DPP-IV and P. horikoshii AAP, bulky regions (gray mesh) hinder simple hydrolase/hydrolase β-sheet formation, and both domains are involved in covering the sticky β-edge. As a consequence, domain positions are fixed. In these enzymes, permanent pores or channel system ensure size selection of substrates.
In monomeric oligopeptidases POP and OPB, the sticky β-strand is covered by a 72- and 95-residue long N-terminal extension, respectively (Fig. 3A (left) and supplemental Fig. 4). Although the extensions join to the amino acids of the propellers in sequence, they run alongside the hydrolase domains and form a complex strap around it. Numerous hydrogen bonds are established between them, the majority of which fix the strap to the sticky β-strand and the two α-helices that stand on either side of it (12–14 and 17 in total, 8–11 and nine to the α-helix/β-strand/α-helix triad in POPs from different sources and in OPB, respectively). Because the structure of POP was determined in the open conformation too, it could be ascertained that these interactions are unchanged by the opening. The N-terminal strap might also be a promoter of monomer stability, but our MD simulations showed that the PhAAP monomer, devoid of such a segment, preserved its structure just as well as POP, supporting the notion that the N-terminal extension might instead be a safeguard against aggregation.
In ApAAP, DPP-IV, and PhAAP, the edge β-strand is unprotected in the monomer; these oligopeptidases multimerize. In ApAAP, the N-terminal extension is only 22 amino acids long, it forms a helix that runs parallel to the α-helix/β-strand/α-helix triad and leaves the sticky β-strand fully unprotected (Fig. 3A, right). However, in the functional ApAAP dimer, the β-strand of one monomer is covered by the corresponding strand of the other in an antiparallel manner, joining their central eight β-strands to form a 16-pleated continuous β-sheet (supplemental Fig. 4). This arrangement results in the two active sites facing in the opposite direction, which allows them to function independently; they might even adopt strikingly different conformations, as was seen in the open/closed mixed dimer structure of ApAAP, where one monomer was found in an open and the other in a closed conformation. Flexibility is retained because only the hydrolase domains participate in the dimerization, and this is essential, because the structure of the closed form of the ApAAP monomer has no side opening to allow access of substrates.
Small hindrances obstruct full access to the exposed β-strands of DPP-IV and PhAAP (Fig. 3B). In DPP-IV, a 30-amino acid-long insertion of blade 4 of the propeller is what reaches to the top of the edge-β-strand, whereas in PhAAP, a 12-residue-long C-terminal extension comes into proximity to the bottom end of it. Both behave as a peg, making it impossible for the head-on dimerization seen in the case of ApAAP to take place. DPP-IV dimerizes; in the dimer, the insertions from each monomer form an X-shaped cross, whereas the eight-pleated central β-sheets meet in a perpendicular orientation. In PhAAP, the loop insertion of blade 3 of one monomer lines up to the sticky β-strand of the other and forms a ninth, antiparallel strand of the central β-sheet making a dimer, whereas the dimers are further organized into the hexamer (supplemental Fig. 4). Because in both DPP-IV and PhAAP, both the hydrolase and the propeller domains participate in multimerization, domain movements necessarily become restricted; thus, these enzymes must possess a fixed substrate entrance for effectiveness, which they do. The mode of multimerization also leads to self-compartmentalizing, especially in the hexamer structure of PhAAP, because the active site openings point inward toward a common central chamber formed by the association. This therefore provides for a sufficient substrate screening mechanism, for maintaining selectivity.
Experimental evidence also supports the suggested role of the N-terminal pegs of oligopeptidases. Removal of the N-terminal segment of POP (where no additional protection for the sticky-β-strand is available) results in aggregation and inactivation depending on the size of the removed segment (52). In contrast, removing the N-terminal extension of ApAAP causes only some destabilization of the enzyme lowering the melting temperature by about 20 K (53).
The eighth β-strand of the central β-sheet supporting the His-loop of α/β-hydrolases is not amylogenic in all cases. Dehalogenases, haloperoxidases, epoxide hydrolases, and hydroxynitrile lyase remove the aggregational risk by inserting prolines (PDB code 2had (54)) or charged amino acids (PDB codes 4fwb,7 3a2m (56), 1a88 (57), 1a7u (57), 3koo (58), 2zjf (59), and 3c70 (60)) into the strand holding the His-loop, lowering their amylogenic propensity considerably, or by introducing a significant twist to this segment (PDB code 2bfn) (61). The sticky β-edge is buried by further, non-amylogenic β-strands in other α/β-hydrolases like acetylcolinesterases, serine carboxypeptidases, and lipases. Peroxisomal α/β-hydrolase Lpx1 forms a dimer in a similar fashion to ApAAP, covering its unprotected amylogenic β-strand by the process (62). However, surprisingly, several monomeric α/β-hydrolases (prolyl iminopeptidase (PDB codes 1qtr (63) and 1azw (64)), thioesterase (PDB code 3gro),8 dienelactone hydrolase (PDB code 1din (55)), and carboxylestherase (PDB code 1auo (5)) provide no protection for their highly amylogenic β-edge (with WALTZ values (45) between 82.4 and 92.2). This might be because other interaction partners, cofactors, glycosylation, or the closeness of the membrane obstructs free access to the implicated β-edge in these enzymes.
Outside the α/β-hydrolase clan, among self-compartmentalizing proteases, tricorn protease, for example, has a multidomain structure similar to that of PhAAP. This enzyme carries out the degradation of small (7–9-amino acid-long) peptides and, just like PhAAP, is also hexameric, although its multimer architecture is of a bit simpler, toroidal shape. Its monomer, however, is more complex than that of PhAAP, consisting of five subdomains. Its α/β catalytic region is capped on both sides by propeller domains; substrates reach the active site by passing the great opening of the toroid and then proceeding through the channel of a seven-bladed propeller, a permanently open route to its active site. Instead of a catalytic triad, a four-residue hydrogen-bonded system of Ser-His-Ser-Glu performs the catalytic reaction. Glu1023 is situated on a long loop connecting an unprotected strand and a distant helix. The seven-bladed propeller is close enough to obstruct dimerization with the corresponding strand of another monomer but too far for shielding; thus, a β-turn-β type insert from the neighboring monomer covers it via hexamerization similarly to that seen in the case of PhAAP.
The proteasome, a further self-compartmentalizing system, forms an immense multilayer, barrel-shaped complex, where four rings, each made up of seven monomers, stack on each other to compose two antechambers and one central catalytic chamber. Proteasome monomers do not have a propeller domain (so they have a very large permanent hole above the hydrolase). The catalytically active Thr is located on the penultimate β-strand of a five-membered sheet, where, in the heptamer, the β-edge of the sheet in one monomer is covered by an insert of the neighboring monomer, resulting in a self-assembled compartment system of inwardly turned active sites in this case too.
Based on the above, we propose that the mode of multimerization and self-assembly among oligopeptidases is fine-tuned by a shielding intent of a sticky β-edge, insertions, and N- and C-terminal extensions, whereas to maintain the effectiveness of catalysis and selectivity, pliability of the His-loop and the position of the propeller blades are adjusted (as summarized in Table 2).
TABLE 2.
Strategies of size selectivity and ways of shielding the sticky β-edge of the hydrolase domain in the prolyl oligopeptidase family
| β-Edge coverage in the monomer | Functional unit | Permanent side entrance | Pliability of His-loop | Substrate screening strategy | Known structures |
|---|---|---|---|---|---|
| Full | Monomer | No | Pliable | Dynamic: inactive open/active closed states | POP, OPBa |
| None | Dimer | No | Pliable | Dynamic: inactive open/active closed states | ApAAP |
| Partial | Self-assembly | Yes | Rigid | Permanent pores or self-compartmentalization | DPP-IV, PhAAP |
a OPB is monomeric; however, no experimental evidence has yet been obtained confirming its ability to open.
Acknowledgments
We acknowledge the ESRF (Grenoble, France) and the EMBL outstation at DESY (Hamburg, Germany) for provision of synchrotron radiation facilities, and we thank Dr. Hassan Belrhali and Dr. Santosh Panjikar for assistance in using beamlines ESRF BM14 and DESY X11, respectively.
This work was supported by Hungarian Scientific Research Fund (OTKA) Grants PD101095, NK101072, and NK67800.

This article contains supplemental Tables 1 and 2 and Figs. 1–5.
The atomic coordinates and structure factors (codes 4HXE, 4HXF, and 4HXG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
T. K. Chiu and M. Li, manuscript in preparation.
M. Lahoda, A. Stsiapanava, J. Mesters, R. Chaloupkova, J. Damborsky, and I. Kuta Smatanova, manuscript in preparation.
E. Dobrovetsky, A. Seitova, Y. Tong, W. Tempel, A. Dong, C. H. Arrowsmith, A. M. Edwards, C. Bountra, J. Weigelt, A. Bochkarev, D. Cossar, and H. Park, manuscript in preparation.
- POP
- prolyl oligopeptidase
- AAP
- acylaminoacyl peptidase
- ApAAP
- A. pernix acylaminoacyl peptidase
- CMK
- benzyloxycarbonyl-glycyl-glycyl-phenylalanyl-chloromethyl ketone
- DPP-IV
- dipeptidyl peptidase IV
- MD
- molecular dynamics
- OPB
- oligopeptidase B
- PhAAP
- P. horikoshii acylaminoacyl peptidase
- PDB
- Protein Data Bank
- TLB
- trilobed protease.
REFERENCES
- 1. Szeltner Z., Polgár L. (2008) Structure, function and biological relevance of prolyl oligopeptidase. Curr. Protein Pept. Sci. 9, 96–107 [DOI] [PubMed] [Google Scholar]
- 2. Polgár L. (2002) The prolyl oligopeptidase family. Cell Mol. Life Sci. 59, 349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fülöp V., Böcskei Z., Polgár L. (1998) Prolyl oligopeptidase. An unusual β-propeller domain regulates proteolysis. Cell 94, 161–170 [DOI] [PubMed] [Google Scholar]
- 4. Szeltner Z., Rea D., Renner V., Fulop V., Polgar L. (2002) Electrostatic effects and binding determinants in the catalysis of prolyl oligopeptidase. Site-specific mutagenesis at the oxyanion binding site. J. Biol. Chem. 277, 42613–42622 [DOI] [PubMed] [Google Scholar]
- 5. Kim K. K., Song H. K., Shin D. H., Hwang K. Y., Choe S., Yoo O. J., Suh S. W. (1997) Crystal structure of carboxylesterase from Pseudomonas fluorescens, an α/β hydrolase with broad substrate specificity. Structure 5, 1571–1584 [DOI] [PubMed] [Google Scholar]
- 6. Li M., Chen C., Davies D. R., Chiu T. K. (2010) Induced-fit mechanism for prolyl endopeptidase. J. Biol. Chem. 285, 21487–21495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shan L., Mathews I. I., Khosla C. (2005) Structural and mechanistic analysis of two prolyl endopeptidases. Role of interdomain dynamics in catalysis and specificity. Proc. Natl. Acad. Sci. U.S.A. 102, 3599–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harmat V., Domokos K, Menyhárd DK, Palló A., Szeltner Z., Szamosi I., Beke-Somfai T., Náray-Szabó G., Polgár L. (2011) Structure and catalysis of acylaminoacyl peptidase. Closed and open subunits of a dimer oligopeptidase. J. Biol. Chem. 286, 1987–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engel M., Hoffmann T., Wagner L., Wermann M., Heiser U., Kiefersauer R., Huber R., Bode W., Demuth H. U., Brandstetter H. (2003) The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. U.S.A. 100, 5063–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engel M., Hoffmann T., Manhart S., Heiser U., Chambre S., Huber R., Demuth H. U., Bode W. (2006) Rigidity and flexibility of dipeptidyl peptidase IV. Crystal structures of and docking experiments with DPIV. J. Mol. Biol. 355, 768–783 [DOI] [PubMed] [Google Scholar]
- 11. Hiramatsu H., Kyono K., Higashiyama Y., Fukushima C., Shima H., Sugiyama S., Inaka K., Yamamoto A., Shimizu R. (2003) The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed β-propeller fold. Biochem. Biophys. Res. Commun. 302, 849–854 [DOI] [PubMed] [Google Scholar]
- 12. Aertgeerts K., Ye S., Tennant M. G., Kraus M. L., Rogers J., Sang B. C., Skene R. J., Webb D. R., Prasad G. S. (2004) Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Sci. 13, 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Szeltner Z., Kiss A. L., Domokos K., Harmat V., Náray-Szabó G., Polgár L. (2009) Characterization of a novel acylaminoacyl peptidase with hexameric structure and endopeptidase activity. Biochim. Biophys. Acta 1794, 1204–1210 [DOI] [PubMed] [Google Scholar]
- 14. Joshua-Tor L., Xu H. E., Johnston S. A., Rees D. C. (1995) Crystal structure of a conserved protease that binds DNA. The bleomycin hydrolase, Gal6. Science 269, 945–950 [DOI] [PubMed] [Google Scholar]
- 15. Kim J. S., Groll M., Musiol H. J., Behrendt R., Kaiser M., Moroder L., Huber R., Brandstetter H. (2002) Navigation inside a protease. Substrate selection and product exit in the tricorn protease from Thermoplasma acidophilum. J. Mol. Biol. 324, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 16. Cha S. S., An Y. J., Lee C. R., Lee H. S., Kim Y. G., Kim S. J., Kwon K. K., De Donatis G. M., Lee J. H., Maurizi M. R., Kang S. G. (2010) Crystal structure of Lon protease. Molecular architecture of gated entry to a sequestered degradation chamber. EMBO J. 29, 3520–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kiss A. L., Hornung B., Rádi K., Gengeliczki Z., Sztáray B., Juhász T., Szeltner Z., Harmat V., Polgár L. (2007) The acylaminoacyl peptidase from Aeropyrum pernix K1 thought to be an exopeptidase displays endopeptidase activity. J. Mol. Biol. 368, 509–520 [DOI] [PubMed] [Google Scholar]
- 18. Palmieri G., Bergamo P., Luini A., Ruvo M., Gogliettino M., Langella E., Saviano M., Hegde R. N., Sandomenico A., Rossi M. (2011) Acylpeptide hydrolase inhibition as targeted strategy to induce proteasomal down-regulation. PLoS One 6, e25888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Naylor S. L., Marshall A., Hensel C., Martinez P. F., Holley B., Sakaguchi A. Y. (1989) The DNF15S2 locus at 3p21 is transcribed in normal lung and small cell lung cancer. Genomics 4, 355–361 [DOI] [PubMed] [Google Scholar]
- 20. Scaloni A., Jones W., Pospischil M., Sassa S., Schneewind O., Popowicz A. M., Bossa F., Graziano S. L., Manning J. M. (1992) Deficiency of acylpeptide hydrolase in small-cell lung carcinoma cell lines. J. Lab. Clin. Med. 120, 546–552 [PubMed] [Google Scholar]
- 21. Erlandsson R., Boldog F., Persson B., Zabarovsky E. R., Allikmets R. L., Sümegi J., Klein G., Jörnvall H. (1991) The gene from the short arm of chromosome 3, at D3F15S2, frequently deleted in renal cell carcinoma, encodes acylpeptide hydrolase. Oncogene 6, 1293–1295 [PubMed] [Google Scholar]
- 22. Duysen E. G., Li B., Xie W., Schopfer L. M., Anderson R. S., Broomfield C. A, Lockridge O. (2001) Evidence for nonacetylcholinesterase targets of organophosphorus nerve agent. Supersensitivity of acetylcholinesterase knockout mouse to VX lethality. J. Pharmacol. Exp. Ther. 299, 528–535 [PubMed] [Google Scholar]
- 23. Richards P. G., Johnson M. K., Ray D. E. (2000) Identification of acylpeptide hydrolase as a sensitive site for reaction with organophosphorus compounds and a potential target for cognitive enhancing drugs. Mol. Pharmacol. 58, 577–583 [DOI] [PubMed] [Google Scholar]
- 24. Kabsch W. (2010) XDS. Acta Crystallogr. D 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 26. Otwinowski Z. (1991) in Proceedings of the CCP4 Study Weekend (Wolf W., Evans P. R., Leslie A. G. W., eds) pp. 80–86, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 27. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 28. Cowtan K. (2006) The Buccaneer software for automated model building. Acta Crystallogr. D 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 29. Vagin A., Teplyakov A. (1997) MOLREP. An automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaguine A. A., Richelle J., Wodak S. J. (1999) SFCHECK. A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 34. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4. Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 36. Mackerell A. D., Jr., Feig M., Brooks C. L., 3rd (2004) Extending the treatment of backbone energetics in protein force fields. Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 [DOI] [PubMed] [Google Scholar]
- 37. Bjelkmar P., Larsson P., Cuendet M. A., Hess B., Lindahl E. (2010) Implementation of the CHARMM force field in GROMACS. Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 6, 459–466 [DOI] [PubMed] [Google Scholar]
- 38. Bosch J., Tamura T., Tamura N., Baumeister W., Essen L. O. (2007) The β-propeller domain of the trilobed protease from Pyrococcus furiosus reveals an open Velcro topology. Acta Crystallogr. D 63, 179–187 [DOI] [PubMed] [Google Scholar]
- 39. Shan L., Mathews I. I., Khosla C. (2005) Structural and mechanistic analysis of two endopeptidases. Role of interdomain dynamics in catalysis and specifity. Proc. Natl. Acad. Sci. U.S.A. 102, 3599–30604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McLuskey K., Paterson N. G., Bland N. D., Isaacs N. W., Mottram J. C. (2010) Crystal stucture of Leishmania major oligopeptidase B gives insight into the enzymatic properties of a trypanosomatid virulence factor. J. Biol. Chem. 285, 39249–39259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kiss A. L., Palló A., Náray-Szabó G., Harmat V., Polgár L. (2008) Structural and kinetic contributions of the oxyanion binding site to the catalytic activity of acylaminoacyl peptidase. J. Struct. Biol. 162, 312–323 [DOI] [PubMed] [Google Scholar]
- 42. Fuxreiter M., Magyar C., Juhász T., Szeltner Z., Polgár L., Simon I. (2005) Flexibility of prolyl oligopeptidase. Molecular dynamics and molecular framework analysis of the potential substrate pathways. Proteins 60, 504–512 [DOI] [PubMed] [Google Scholar]
- 43. Goettig P., Groll M., Kim J. S., Huber R., Brandstetter H. (2002) Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism. EMBO J. 21, 5343–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richardson J. S., Richardson D. C. (2002) Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. U.S.A. 99, 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maurer-Stroh S., Debulpaep M., Kuemmerer N., Lopez de la Paz M., Martins I. C., Reumers J., Morris K. L., Copland A., Serpell L., Serrano L., Schymkowitz J. W., Rousseau F. (2010) Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods 7, 237–242 [DOI] [PubMed] [Google Scholar]
- 46. Dobson C. M. (2003) Protein folding and misfolding. Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 47. Clark L. A. (2005) Protein aggregation determinants from a simplified model. Cooperative folders resist aggregation. Protein Sci. 14, 653–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Monsellier E., Chiti F. (2007) Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 8, 737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Soldi G., Bemporad F., Torrassa S., Relini A., Ramazzotti M., Taddei N., Chiti F. (2005) Amyloid formation of a protein in the absence of initial unfolding and destabilization of the native state. Biophys. J. 89, 4234–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chiti F., Dobson C. M. (2009) Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 5, 15–22 [DOI] [PubMed] [Google Scholar]
- 51. Stefani M., (2004) Protein misfolding and aggregation. New examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta 1739, 5–25 [DOI] [PubMed] [Google Scholar]
- 52. Juhász T., Szeltner Z., Polgár L. (2007) Truncated prolyl oligopeptidase from Pyrococcus furiosus. Proteins 69, 633–643 [DOI] [PubMed] [Google Scholar]
- 53. Zhang Z., Zheng B., Wang Y., Chen Y., Manco G., Feng Y. (2008) The conserved N-terminal helix of acylpeptide hydrolase from archaeon Aeropyrum pernix K1 is important for its hyperthermophilic activity. Biochim. Biophys. Acta 1784, 1176–1183 [DOI] [PubMed] [Google Scholar]
- 54. Franken S. M., Rozeboom H. J., Kalk K. H., Dijkstra B. W. (1991) Crystal structure of haloalkane dehalogenase. An enzyme to detoxify halogenated alkanes. EMBO J. 10, 1297–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pathak D., Ollis D. (1990) Refined structure of dienelactone hydrolase at 1.8 Å. J. Mol. Biol. 214, 497–525 [DOI] [PubMed] [Google Scholar]
- 56. Prokop Z., Sato Y., Brezovsky J., Mozga T., Chaloupkova R., Koudelakova T., Jerabek P., Stepankova V., Natsume R., van Leeuwen J. G., Janssen D. B., Florian J., Nagata Y., Senda T., Damborsky J. (2010) Enantioselectivity of haloalkane dehalogenases and its modulation by surface loop engineering. Angew. Chem. Int. Ed. Engl. 49, 6111–6115 [DOI] [PubMed] [Google Scholar]
- 57. Hofmann B., Tölzer S., Pelletier I., Altenbuchner J., van Pée K. H., Hecht H. J. (1998) Structural investigation of the cofactor-free chloroperoxidases. J. Mol. Biol. 279, 889–900 [DOI] [PubMed] [Google Scholar]
- 58. Eldrup A. B., Soleymanzadeh F., Farrow N. A., Kukulka A., De Lombaert S. (2010) Optimization of piperidyl-ureas as inhibitors of soluble epoxide hydrolase. Bioorg. Med. Chem. Lett. 20, 571–575 [DOI] [PubMed] [Google Scholar]
- 59. Biswal B. K., Morisseau C., Garen G., Cherney M. M., Garen C., Niu C., Hammock B. D., James M. N. (2008) The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J. Mol. Biol. 381, 897–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schmidt A., Gruber K., Kratky C., Lamzin V. S. (2008) Atomic resolution crystal structures and quantum chemistry meet to reveal subtleties of hydroxynitrile lyase catalysis. J. Biol. Chem. 283, 21827–21836 [DOI] [PubMed] [Google Scholar]
- 61. Monincová M., Prokop Z., Vévodová J., Nagata Y., Damborsky J. (2007) Weak activity of haloalkane dehalogenase LinB with 1,2,3-trichloropropane revealed by x-ray crystallography and microcalorimetry. Appl. Environ. Microbiol. 73, 2005–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thoms S., Hofhuis J., Thöing C., Gärtner J., Niemann H. H. (2011) The unusual extended C-terminal helix of the peroxisomal α/β-hydrolase Lpx1 is involved in dimer contacts but dispensable for dimerization. J. Struct. Biol. 175, 362–371 [DOI] [PubMed] [Google Scholar]
- 63. Rould M. A., Perona J. J., Steitz T. A. (1991) Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature 352, 213–218 [DOI] [PubMed] [Google Scholar]
- 64. Medrano F. J., Alonso J., García J. L., Romero A., Bode W., Gomis-Rüth F. X. (1998) Structure of proline iminopeptidase from Xanthomonas campestris pv. citri. A prototype for the prolyl oligopeptidase family. EMBO J. 17, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]