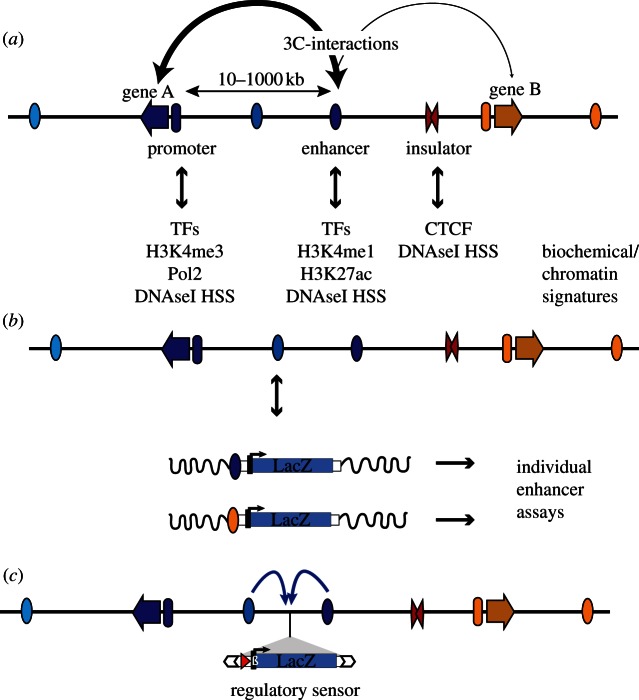
Figure 1.
Regulatory elements and how to detect them. (a) Multiple regulatory elements are involved in the tissue-specific transcriptional regulation of genes (arrows). These elements include promoters (ovals preceding genes), enhancers (ovals, with tissue-specific activity indicated by different colours) and insulators (red double triangle). Enhancers can be located at great distance from their target gene. Different regulatory elements are marked by distinct chromatin signatures and binding of proteins, which can be used to identify active elements in accessible tissues using biochemical methods. It is generally accepted that activation is achieved through direct interactions between enhancers and target promoters, which can be detected by chromosome conformation capture (3C) techniques (the stroke of the connecting arrows represents the frequency/strength of the interactions). TFs, transcription factors; DNaseI HSS, DNaseI hypersensitive site. (b) The activity of individual regulatory modules can be tested in in vivo reporter assays. This is usually achieved by cloning the element upstream of a reporter gene, by measuring the expression of this transgene integrated randomly into the genome (indicated by wavy black line). (c) In comparison, GROMIT uses a transposon (white arrows), which carries regulatory sensor (LacZ gene, driven by minimal promoter) as cargo. Through random transposition, the reporter gene can be distributed throughout the genome, showing integrated regulatory input from the multiple modules (indicated by arrows) acting on that position.