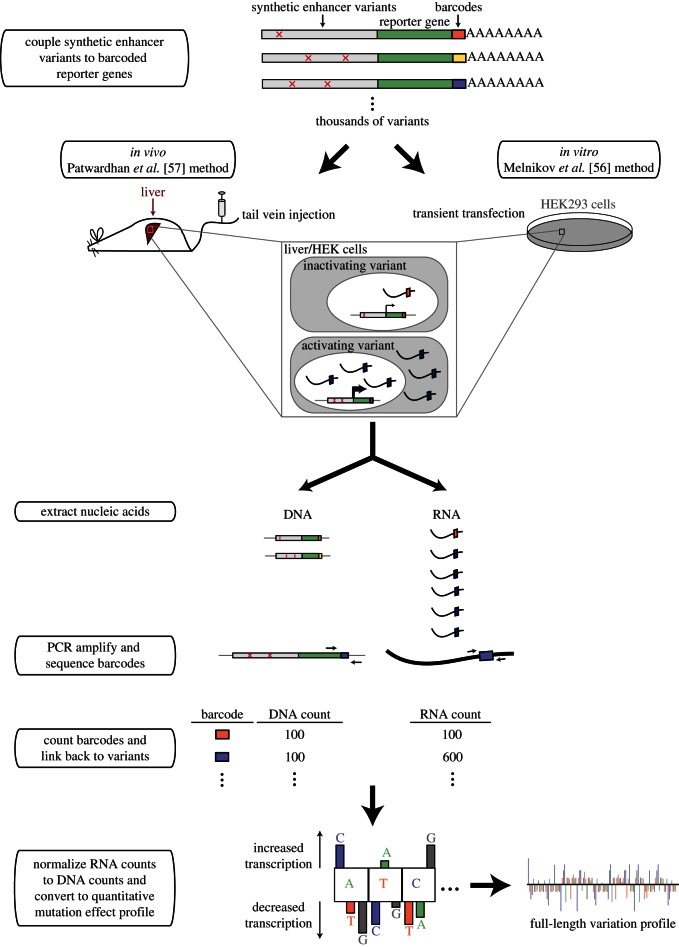
Figure 2.
High-throughput methods to quantitatively assess enhancer variation. In both methods discussed, enhancer variants and unique barcodes are synthesized and linked to reporter genes. Enhancer–reporter constructs are then delivered in vivo to liver cells through mouse tail vein injection (Patwardhan et al. [57]) or by transient transfection to human HEK293T cells (Melnikov et al. [56]). RNA and DNA are collected, and the barcodes contained in each nucleic acid fraction are PCR amplified and sequenced. The number of RNA sequence reads for each barcode is normalized by dividing by its number of DNA reads to create a measure of reporter gene expression. Comparing reporter gene expression from enhancer variants to the wild-type enhancer results in a quantitative mutation effect profile.