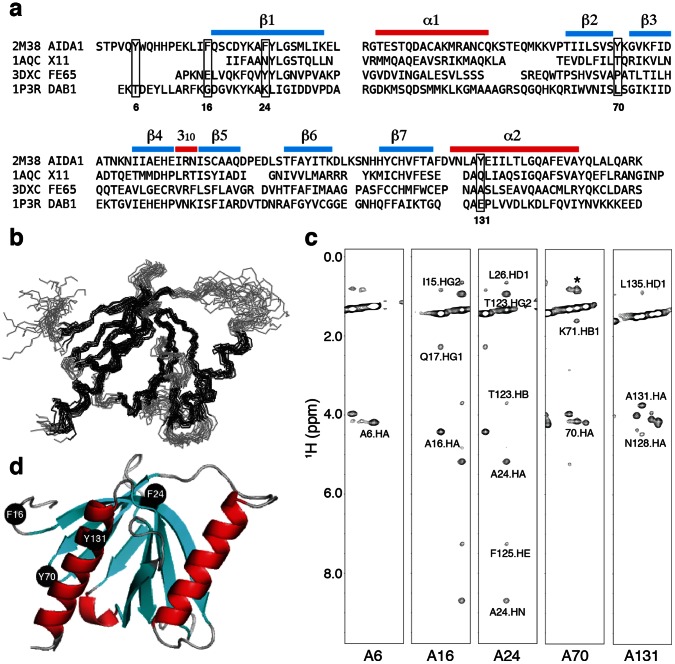
Figure 1. (a) Sequence alignment of the AIDA1 PTB domain against the APP binding proteins, Dab1 [25], X11 [17] and Fe65 [20].
Five aromatic amino acids selected for alanine substitution in AIDA1 PTB domain are boxed. (b) Backbone atom superposition of top15 structures according to lowest refinement energy. (c) Strip plots of a 13C-edited NOESY spectrum at the Cβ chemical shift of each alanine substituted in the PTB5M mutant. A asterisk denotes a resonance not associated with that strip. (d) A ribbon representation of the PTB5M model highlighting the positions of the alanine substitutions. Y6A is not shown in the figure as the first 14 amino acids are unstructured and were excluded from the structure calculation.