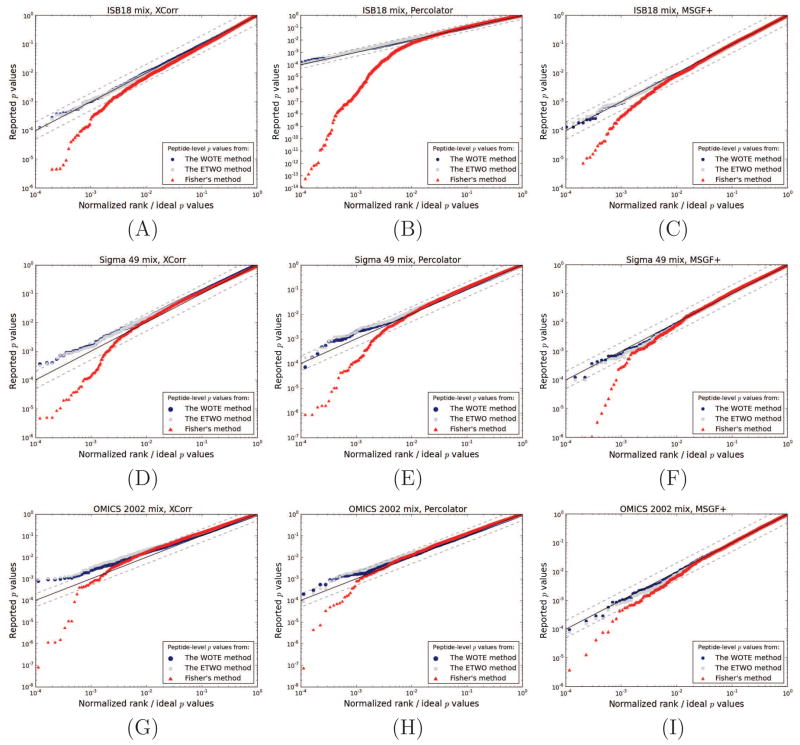
Figure 2. The calibration of peptide-level p values from WOTE, ETWO and Fisher’s method.
Three different datasets of known protein mixtures were scored against a bipartite target and a reversed decoy database using Crux, MSGF+ and Percolator. Subsequently, p values were estimated using either the SEQUEST’s XCorr (left side panels), the Percolator score (middle panels) or scores from MSGF+ (right side panels). Entrapment peptide p values are plotted relative to an ideal, uniform distribution of p values. The y = x diagonal is indicated by a black line, and y = 2x and y = x/2 are shown by dashed lines. Panels (A), (B) and (C) show results from the ISB18 mix. Panels (D), (E) and (F) represent the Sigma 49 mix, and panels (G), (H) and (I) represent the OMICS 2002 mix.