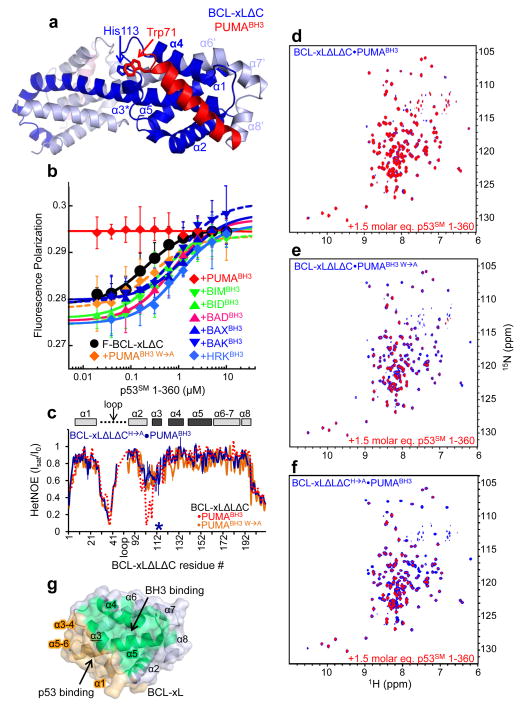
Figure 2.
Mechanism of PUMA binding-induced p53 release from BCL-xL. a. Crystal structure of BCL-xLΔC domain-swapped dimer bound to PUMABH3. The two subunits of BCL-xLΔC are colored dark blue and light blue, respectively, and the eight α-helices of one globular core of the dimer (right side) are labeled α1-α5 and α6′-α8′. The two molecules of PUMABH3 are colored red (front, right) and light red (back, left), respectively. The imidazole ring of His113 of BCL-xL and the indole ring of Trp71 of PUMABH3 that are engaged in a π-stacking interaction are indicated. b. Fluorescence polarization analysis of titrations of p53SM 1-360 into fluorescently labeled BCL-xLΔC (F-BCL-xLΔC), isolated or previously bound to a slight molar excess of different BH3 peptides from PUMA, BIM, BID, BAD, BAX, BAK or HRK as indicated. Error bars represent the standard error of the mean of five independent titrations. c. Sequence dependence of {1H}-15N HetNOE values for BCL-xLΔLΔC in complex with PUMABH3 W→A (orange) and BCL-xLΔLΔCH→A in complex with wild-type PUMABH3 (blue; the mutation site is marked with an asterisk). The values observed for BCL-xLΔLΔC·PUMABH3 (Fig. 1g) are illustrated with a dashed red line. d–f. Overlaid 2D 1H-15N TROSY spectra of 100 μM 15N-BCL-xLΔLΔC bound to unlabeled PUMABH3 (d), PUMABH3 W→A (e) or 15N-BCL-xLΔLΔCH→A bound to PUMABH3 (f) in the absence (blue) and presence (red) of a 1.5 molar excess of p53SM 1-360. g. Surface representation of apo BCL-xL highlighting the non-overlapping nature of its surfaces that bind to BH3 domains, including PUMABH3 (green) and p53 (orange).