Abstract
Long after their original discovery, termite gut spirochetes were recently isolated in pure culture for the first time. They revealed metabolic capabilities hitherto unknown in the Spirochaetes division of the Bacteria, i.e., H2 plus CO2 acetogenesis (J. R. Leadbetter, T. M. Schmidt, J. R. Graber, and J. A. Breznak, Science 283:686-689, 1999) and dinitrogen fixation (T. G. Lilburn, K. S. Kim, N. E. Ostrom, K. R. Byzek, J. R. Leadbetter, and J. A. Breznak, Science 292:2495-2498, 2001). However, application of specific epithets to the strains isolated (Treponema strains ZAS-1, ZAS-2, and ZAS-9) was postponed pending a more complete characterization of their phenotypic properties. Here we describe the major properties of strain ZAS-9, which is readily distinguished from strains ZAS-1 and ZAS-2 by its shorter mean cell wavelength or body pitch (1.1 versus 2.3 μm), by its nonhomoacetogenic fermentation of carbohydrates to acetate, ethanol, H2, and CO2, and by 7 to 8% dissimilarity between its 16S rRNA sequence and those of ZAS-1 and ZAS-2. Strain ZAS-9 is proposed as the type strain of the new species, Treponema azotonutricium. Strains ZAS-1 and ZAS-2, which are H2-consuming, CO2-reducing homoacetogens, are proposed here to be two strains of the new species Treponema primitia. Apart from the salient differences mentioned above, the genomes of all three strains were similar in size (3,461 to 3,901 kb), in G+C content (50.0 to 51.0 mol%), and in possession of 2 copies of the gene encoding 16S rRNA (rrs). For comparison, the genome of the free-living spirochete Spirochaeta aurantia strain J1 was analyzed by the same methods and found to have a size of 3,719 kb, to contain 65.6 mol% G+C, and also to possess 2 copies of the rrs gene.
The conspicuous abundance of spirochetes in termite hindguts was noted by Leidy more than 100 years ago (20, 21), and to this day termites remain an ideal source material for classroom demonstration of the morphological diversity and motility of spirochetes (1, 8). Nevertheless, our understanding of termite hindgut spirochetes and their importance to termite vitality has long been meager, mainly because none had ever been isolated and studied in vitro. Recently, this situation has changed dramatically. Cultivation-independent (molecular phylogenetic) analysis of several hundred spirochetal 16S rRNA gene clones obtained from a variety of termites revealed that all were affiliated with the genus Treponema. However, the termite-derived clones were not closely related to other known treponemes or to treponemal 16S rRNA gene clones obtained from other animals. Almost all of them assembled into a distinct subgroup—the “termite cluster”—within the genus. Moreover, the clones themselves were remarkably diverse at what could be considered the species level (i.e., many had 16S rRNA sequences ≤97% similar to each other [28]), even within individual termites, which were found to harbor two dozen or more species of Treponema (5, 23).
Close on the heels of the phylogenetic analyses, we reported the isolation of the first pure cultures after many years of periodic but unsuccessful attempts in our laboratory. This achievement revealed that termite gut treponemes possess two metabolic capabilities hitherto unknown within the Spirochaetes division of the Bacteria, i.e., H2-CO2 acetogenesis (19) and dinitrogen fixation (22). Both properties were relevant to the association of spirochetes with their host: microbially produced acetate is an important carbon and energy source for termites, and gut microbe-mediated dinitrogen fixation is an important source of N for termite growth (4, 5). Accordingly, the first experiments were focused on these metabolic processes. The isolates were simply referred to as Treponema strains ZAS-1, ZAS-2, and ZAS-9, with assignment of specific epithets postponed until a more detailed characterization of the strains could be performed. This has now been done.
In this paper, we describe the major phenotypic properties of strain ZAS-9 supporting its recognition as the type strain of the new species Treponema azotonutricium. A companion paper (11) presents a detailed characterization of strains ZAS-1 and ZAS-2, which are similar to each other and distinct from strain ZAS-9, and for which we here propose the new species name Treponema primitia. We also provide data on the genomes of ZAS strains (and of a free-living spirochete, Spirochaeta aurantia [6, 7], analyzed for comparison) in terms of molecular size, G+C content, and rrs copy number.
MATERIALS AND METHODS
Organisms, media, and nutritional studies.
Treponema strain ZAS-9 (DSM 13862; ATCC BAA-888) was isolated from a tube in an anoxic agar dilution series that had received the equivalent of 0.001 gut contents from a small (ca. 10 mg [fresh weight]) worker larva of Zootermopsis angusticollis (Hagen) (Isoptera: Termopsidae). A portion of a well-isolated, subsurface colony was picked with a sterile Pasteur pipette and subjected to two successive reisolations in agar dilution series, both of which yielded subsurface colonies that were homogenous in size (ca. 1 to 2 mm), shape (spherical), and color (semitransparent, dull white with a slight orange hue that may have been due to the rifamycin in the medium [see below]) and which often had adjacent cracks in the agar, apparently resulting from gas production. The isolation medium was a rifamycin-, phosphomycin-, and bromoethanesulfonate-containing medium similar to that used for the isolation of strains ZAS-1 and ZAS-2 (19), but it also contained 0.1% (vol/vol) yeast autolysate (19), 2% (vol/vol) cofactor solution (19), 1.3 g of cellobiose/liter, and 0.4 g (each) of d-maltose, d-sucrose, d-trehalose, d-glucose, d-fructose, d-mannose, l-rhamnose, d-mannitol, d-arabinose, l-arabinose, l-fucose, d-ribose, d-xylose, sodium d-glucuronate, and sodium d-galacturonate/liter, and it was solidified by incorporating 0.8% Ultrapure agarose (Gibco). Incubation was carried out at 23°C under an atmosphere of 80% H2 (the balance was CO2). After isolation, strain ZAS-9 was routinely grown under 20% CO2 (the balance was N2, or H2 when H2 was tested as an energy source) in a dithiothreitol-reduced mineral medium supplemented with trace elements, vitamins, cofactors, and 2% (vol/vol) yeast autolysate (2YACo medium [11, 19]), as well as with a disaccharide mixture (cellobiose, maltose, sucrose, and trehalose at 5 mM each). Culture tubes (butyl rubber-stoppered anaerobe tubes containing 5 ml of liquid plus ca. 21 ml of the gas phase) were placed horizontally on a reciprocating shaker (ca. 100 strokes/min), and turbidimetric measurements and direct cell counts were performed as previously described (22). Utilization of various individual substrates as energy sources was examined by using 2YACo basal medium without the disaccharide mixture.
Strains ZAS-1 (DSM 12426) and ZAS-2 (DSM 12427; ATCC BAA-887) were isolated from enrichment cultures as previously described (19) and grown under H2-CO2 (80:20, vol/vol) in 2YACo medium supplemented with 5 mM maltose (11). S. aurantia strain J1 (ATCC 25082) was grown under air with shaking (120 rpm) in Erlenmeyer flasks containing MPYM medium as one-fourth their volume. MPYM medium contained 0.2% each maltose, peptone, and yeast extract and 10 mM 3-N-(morpholino)propanesulfonic acid (MOPS). The medium was adjusted to pH 7.5 prior to heat sterilization. Unless otherwise indicated, all spirochetes were grown at 30°C. Cells of Myxococcus xanthus strain DK1622 were kindly provided by K. Stredwick.
Metabolic studies.
Fermentation products of strain ZAS-9 were determined after growth of cells in 2YACo basal medium containing a growth-limiting amount of maltose (5 mM), which was assumed to have been completely utilized when growth ceased. H2 and acetate were analyzed by using gas chromatography and high-performance liquid chromatography (HPLC), respectively (19). Ethanol was quantified by enzymatic analysis (332-A Ethanol Assay kit; Sigma-Aldrich, St. Louis, Mo.).
Hydrogenase, formate dehydrogenase, and CO dehydrogenase activities of ZAS-9 were determined by using a suspension of cetyltrimethylammonium bromide (CTAB)-permeabilized cells (19). Cells were derived from a late-exponential-phase culture grown in 2YACo medium supplemented with disaccharides (described above). The presence of catalase was examined by adding a drop of 3% H2O2 to a small portion of a harvested cell pellet on a glass slide, covering the mixture with a coverslip, and observing for production of gas (O2) bubbles.
Analyses of genomic DNA.
Genome sizes were determined by pulsed-field gel electrophoresis (PFGE) of restriction digests of genomic DNA (3). Twelve different rare-cutting restriction enzymes were evaluated, and those yielding the fewest number of well-resolved fragments were selected for use. PFGE was performed by using a CHEF-DR II apparatus (Bio-Rad Laboratories, Richmond, Calif.) and the conditions described by Bergthorsson and Ochman (2), which included varying the pulse times and run duration to resolve fragments within various size ranges. Markers for PFGE included Yeast Chromosome, Lambda Ladder, MidRange II, and Low Range (all from New England Biolabs, Inc., Beverly, Mass.).
G+C content was determined by HPLC analysis of P1 nuclease- and alkaline phosphatase-digested samples as described by Mesbah et al. (24), except that (i) DNA was purified by using a genomic DNA kit with a 55/G purification column (Qiagen Inc., Chatsworth, Calif.) and (ii) the HPLC column used was an Altima C18 column (250 by 4.6 mm; particle size, 5 μm) (Alltech Associates, Deerfield, Ill.).
The rrs gene copy number was determined by Southern hybridizations of restricted DNA with an rrs-targeted probe (15; updates described in reference 17). Escherichia coli genomic DNA was used as a control.
Microscopy.
Phase-contrast micrographs were prepared from wet mounts on agar-coated slides (26). Images were captured by using a Zeiss Axioskop microscope (Carl Zeiss, Inc., Thornwood, N.Y). equipped with a SPOT charge-coupled-device digital camera (Diagnostic Instruments, Inc., Sterling Heights, Mich.). Cells for electron microscopy were prepared as previously described (18). Images were obtained by using a JEOL 100CXII transmission electron microscope (JEOL-USA, Inc., Peabody, Mass.) operated by staff at the Center for Advanced Microscopy at Michigan State University.
RESULTS AND DISCUSSION
Morphological distinction of ZAS strains.
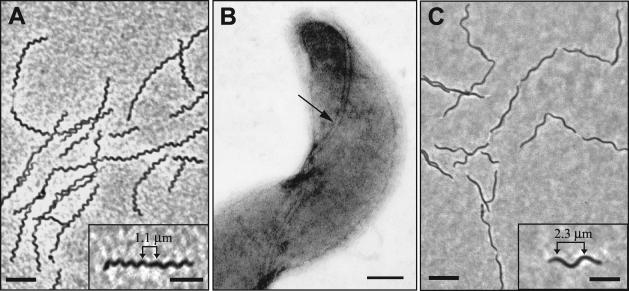
Cells of ZAS-9 measured 0.2 to 0.3 μm wide by 10 to 12 μm long and possessed a wavelength or cell body pitch of 1.1 ± 0.1 μm (n = 15) (Fig. 1A). On this basis, they are readily distinguished from cells of ZAS-1 and ZAS-2, which are similar in width but shorter (3 to 7 μm) and appear more loosely coiled (cell wavelength or body pitch, 2.3 ± 0.2 μm [n = 15]) (Fig. 1C). All three strains, however, possessed two periplasmic flagella, inserted at opposite ends of the cell (Fig. 1B).
FIG. 1.
Morphology of T. azotonutricium strain ZAS-9 (A and B) and T. primitia strain ZAS-2 (C) by phase-contrast (A and C) and transmission electron (B) microscopy. Insets in panels A and C show single cells of each strain, with wavelengths indicated by arrowed bars. One of the periplasmic flagella of ZAS-9 is seen in panel B (arrow). Strain ZAS-1 is morphologically indistinguishable from strain ZAS-2. Bars, 5 μm for panels A and C; 2.5 μm for insets in panels A and C; 0.1 μm for panel B.
Nutrition, growth characteristics, and fermentation products.
Strain ZAS-9 was a strict anaerobe and catalase negative. Substrates utilized as energy sources by strain ZAS-9 were limited to carbohydrates, which supported growth at cell doubling times ranging from 35 to 47 h at 30°C (Table 1). Doubling times were about 1.3 times longer at 23°C, and no growth occurred at 37°C. Unlike strains ZAS-1 and ZAS-2, which are homoacetogens that can grow by CO2-reductive acetogenesis with H2 as the electron donor (11, 19), strain ZAS-9 neither used H2 (plus CO2) as an energy source (Table 1) nor grew faster or to greater cell yields when H2 was included along with a fermentable sugar that would support growth (data not shown). On the contrary, H2 was a major product of strain ZAS-9, which fermented maltose (C12H22O11) to acetate (2.80 mol per mol of maltose fermented), ethanol (0.80 mol/mol), H2 (5.20 mol/mol), and CO2 (3.60 mol/mol; assumed to be equal to acetate plus ethanol). Carbon and electron recoveries were 90 and 88%, respectively, and HPLC profiles of spent medium revealed no other products in significant amounts. In agreement with these observations, enzyme assays revealed that strain ZAS-9 possessed hydrogenase activity (1.15 U/mg of protein) but no detectable formate dehydrogenase or CO dehydrogenase activity. The latter two enzymes are characteristic of the Wood-Ljungdahl pathway for CO2-reductive acetogenesis and are present in strains ZAS-1 and ZAS-2 (19).
TABLE 1.
Substrates used as energy sources by Treponema strain ZAS-9
| Substratea | Doubling time (h) | Yield (108 cells/ml)b |
|---|---|---|
| Glucose | 35 | 3.1 |
| Fructose | 37 | 2.6 |
| Ribose | 42 | 1.4 |
| Xylose | 35 | 2.8 |
| Maltose | 47 | 5.6 |
| Cellobiose | 45 | 4.8 |
| None (control) | No growth |
Substrates were provided at a final concentration of 5 mM, except for H2 (80% of headspace volume; the balance was CO2). Mannitol, arabinose, sucrose, trehalose, glycine, sodium lactate, sodium pyruvate, uric acid, and H2 were not utilized.
1.0 × 108 cells is equivalent to 24.7 μg of protein.
Assuming that protein makes up 55% of dry cell mass (25) in spirochetes, it can be calculated from Table 1 that the yield of ZAS-9 grown on 5 mM maltose (a concentration that was growth-limiting in the otherwise complex, yeast autolysate-containing medium [see Materials and Methods]) was ca. 50.3 g (dry mass)/mol of maltose. If ATP formation during maltose fermentation occurred solely from glycolysis (2 net mol of ATP/mol of glucosyl unit of maltose) plus acetate kinase-mediated acetate formation (1 mol of ATP/mol of acetate formed), then the maltose fermentation balance (described above) implies that cells derive 6.8 net mol of ATP/mol of maltose, giving an apparent YATP value of 7.4 g (dry mass)/mol of ATP. This is at the lower end of YATP values (mean, 10.5 g [dry mass]/mol of ATP) determined for a variety of bacteria grown fermentatively in complex media (30).
One explanation for the seemingly low YATP value may be the relatively large amount of H2 produced during fermentation (equivalent to 2.6 mol of H2/mol of glucosyl unit of maltose; see above). If ZAS-9 ferments the glucosyl units by using the Embden-Meyerhof pathway accompanied by a phosphoroclastic cleavage of pyruvate, as does Spirochaeta stenostrepta (13), one of the closest cultivable phylogenetic relatives of ZAS-9 other than strains ZAS-1 and ZAS-2 (22), then acetate, ethanol, and CO2 arise following the oxidation of pyruvate by pyruvate ferredoxin oxidoreductase (E0′, −0.40 V), a reaction that can readily be coupled to hydrogenase-mediated reduction of protons to H2 (E0′, −0.41 V) and can account for 4.0 of the 5.2 mol of H2 produced per mol of maltose. The remaining H2 (1.2 mol per mol of maltose) would then presumably come from the oxidation of NADH (E0′, −0.32 V), formed during glycolysis via glyceraldehyde-3-phosphate dehydrogenase (phosphorylating; EC 1.2.1.12). However, assuming an intracellular steady-state concentration of 12.56 mM NAD+ and 3.36 mM NADH (31), H2 production from NADH at 30°C becomes thermodynamically unfavorable at ambient H2 concentrations of >2 × 10−4 atm (27). Given the conditions under which the fermentation was carried out (5 ml of culture fermenting 5 mM maltose in a tube with 21 ml of headspace), less than 0.01 mol of H2 per mol of maltose could have arisen from NADH unless its oxidation were coupled to the consumption of energy. Hence, some of the energy conserved by ZAS-9 during fermentation may be invested in the production of H2 from NADH (1.2 mol of H2 per mol of maltose) rather than biomass, resulting in the low apparent YATP.
The mechanism of such putative energy-dependent formation of H2 remains to be elucidated. It is unlikely to involve the intrinsic H2-producing activity of dinitrogenase of strain ZAS-9, which is repressed under these growth conditions (22), but it could conceivably involve a coupling of hydrogenase with dinitrogenase reductase—the ATP-consuming, low-potential electron donor for dinitrogenase. It could also involve reverse electron transport mediated by NADH:ferredoxin oxidoreductase and hydrogenase, as has been found in the swine dysentery spirochete Brachyspira hyodysenteriae, which also produces copious amounts of hydrogen during glucose metabolism (29). In any case, the limited production of biomass by ZAS-9 during sugar fermentation parallels the low biomass yields of the homoacetogenic strains (ZAS-1 and ZAS-2) when grown on glucose or H2 plus CO2, and it is consistent with the notion that these symbiotic spirochetes have been selected for conversion of substrates to products (particularly acetate, a major energy source for termites), not for rapid growth or efficient conversion of substrates into cell material (11). In this regard it is worth noting that, where quantitative analyses have been done, most free-living spirochetes that evolve H2 during carbohydrate fermentation (including S. stenostrepta and Spirochaeta zuelzerae, which group within the genus Treponema on the basis of their 16S rRNA sequences [23]) produce far less H2 than ZAS-9 (≤1.8 mol/mol of glucose) (references 9, 12, and 33 and references therein). An exception is the thermophilic spirochete Spirochaeta thermophila (2.9 mol of H2/mol of glucose) (14).
Genomic analyses.
In an effort to characterize some fundamental properties of the genomes of the ZAS strains, we determined their molecular sizes, G+C contents, and copy numbers of the 16S rRNA-encoding gene (rrs). For comparison, these properties were also determined for the free-living spirochete S. aurantia (6, 7). Results (Table 2; Fig. 2 and 3) revealed that the genomes of the ZAS strains were similar in size (3,461 to 3,901 kb) and G+C content (50.0 to 51.0 mol%) and that each possessed 2 copies of rrs. The genomes of these termite gut treponemes were substantially larger than those of the syphilis agent, Treponema pallidum (1,138 kb) (10), and the human oral treponeme Treponema denticola (ca. 2,800 kb) (32).
TABLE 2.
Properties of genomic DNAs of Treponema strains ZAS-1, ZAS-2, and ZAS-9 and of the free-living spirochete S. aurantia J1
| Strain | Mean genome size (kb)a ± SEMb (restriction enzyme used) | Meanc (range) G+C content (mol%) | rrs copy no.d |
|---|---|---|---|
| T. primitia | |||
| ZAS-1 | 3,461 ± 14 (PmeI) | 51.0 (50.9-51.2) | 2 |
| ZAS-2 | 3,835 ± 24 (XbaI) | 50.9 (50.6-51.1) | 2 |
| T. azotonutricium ZAS-9 | 3,901 ± 47 (PmeI) | 50.0 (49.8-50.1) | 2 |
| S. aurantia J1 | 3,719 ± 13 (AseI) | 65.6 (65.4-65.7) | 2 |
Mean sizes (in kilobases) of genomic fragments resolved by PFGE were as follows (with the number of independent determinations per fragment given in parentheses after each): for strain ZAS-1, 574 (n = 8), 260 (n = 7), 243 (n = 7), 238 (n = 8), 220 (n = 8), 191 (n = 7), 186 (n = 6), 161 (n = 12), 143 (n = 8), 131 (n = 10), 120 (n = 10), 107 (n = 10), 100 (n = 10), 92 (n = 9), 89 (n = 5), 85 (n = 4), 72 (n = 6), 65 (n = 6), 58 (n = 6), 51 (n = 6), 49 (n = 6), 46 (n = 6), 40 (n = 6), 33 (n = 6), 29 (n = 6), 26 (n = 5), 21 (n = 3), 16 (n = 2), and 15 (n = 2); for strain ZAS-2, 326 (n = 6), 237 (n = 9), 225 (n = 5), 217 (n = 9), 213 (n = 5), 197 (n = 8), 175 (n = 7), 165 (n = 8), 160 (n = 4), 134 (n = 9), 133 (n = 2), 127 (n = 9), 121 (n = 11), 113 (n = 11), 105 (n = 7), 99 (n = 6), 95 (n = 4), 91 (n = 4), 87 (n = 4), 78 (n = 4), 64 (n = 6), 62 (n = 2), 60 (n = 2), 59 (n = 5), 54 (n = 5), 49 (n = 5), 46 (n = 5), 43 (n = 5), 42 (n = 2), 35 (n = 2), 32 (n = 2), 30 (n = 2), 28 (n = 5), 27 (n = 2), 26 (n = 2), 23 (n = 3), 22 (n = 3), 20 (n = 3), and 18 (n = 3); for strain ZAS-9, 1,353 (n = 10), 941 (n = 10), 763 (n = 10), 323 (n = 15), 163 (n = 15), 136 (n = 15), 87 (n = 15), 72 (n = 5), and 63 (n = 5); for S. aurantia, 664 (n = 9), 350 (n = 3), 318 (n = 3), 296 (n = 3), 255 (n = 6), 225 (n = 6), 176 (n = 4), 174 (n = 4), 131 (n = 21), 121 (n = 21), 119 (n = 7), 116 (n = 7), 107 (n = 25), 103 (n = 25), 88 (n = 28), 85 (n = 28), 81 (n = 28), 68 (n = 28), 55 (n = 28), 48 (n = 28), 40 (n = 28), 34 (n = 28), 21 (n = 12), 20 (n = 12), 13 (n = 13), and 9 (n = 12).
Standard errors of the means reported are the summations of those calculated for each individual fragment. The average coefficient of variation ranged from 1.1 to 3.0%.
Mean for two independent DNA preparations; three HPLC analyses were performed per preparation. G+C content was calculated from apparent ratios of dGuo to deoxyribosylthymine as previously described (24). The mean G+C content of control DNA from M. xanthus DK1622 (67.4 mol%; range, 67.3 to 67.6 mol%) agreed well with that previously reported (67.55 ± 0.02 mol%) (24).
The restriction enzymes used for this determination were known from 16S rRNA gene sequence data not to cut within the probe target region (positions 8 to 536 by E. coli numbering).
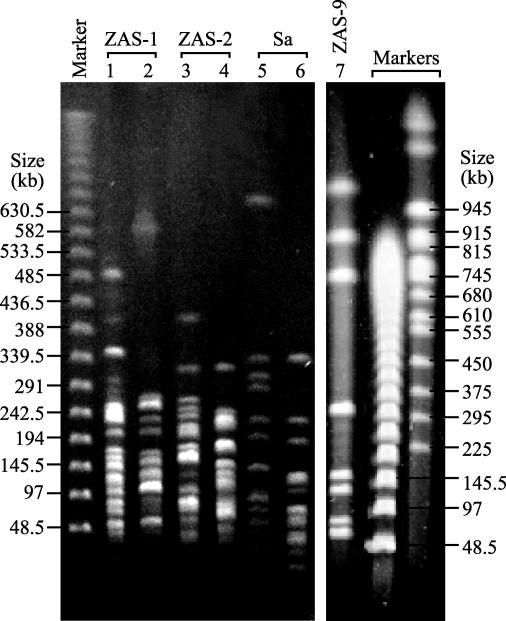
FIG. 2.
Pulsed-field gel electropherograms of digested genomic DNAs of T. primitia strains ZAS-1 and ZAS-2, T. azotonutricium ZAS-9, and S. aurantia J1 (Sa). Restriction enzymes used for digestion were as follows: XbaI (lanes 1 and 4), PmeI (lanes 2 and 7), SwaI (lane 3), AseI (lane 5), and SspI (lane 6).
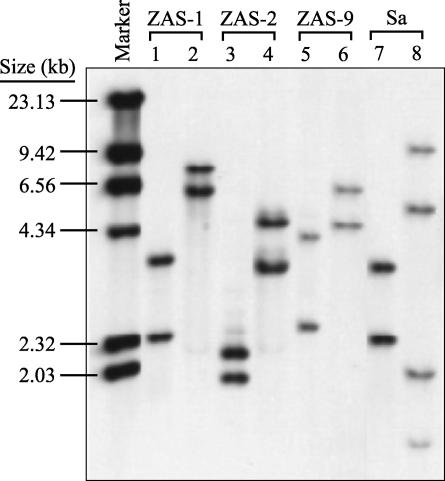
FIG. 3.
Southern hybridizations of digested genomic DNAs of T. primitia strains ZAS-1 and ZAS-2, T. azotonutricium ZAS-9, and S. aurantia J1 (Sa) showing rrs copy number. Restriction enzymes used were SmaI (lanes 1, 3, 5, and 7), KpnI (lanes 2, 4, and 6), and NaeI (lane 8). NaeI is known to cut within rrs of S. aurantia, resulting in four hybridizable fragments (two per gene). The rrs copy number of PvuII-digested E. coli, used as a control, was 7 (data not shown), as previously reported (16).
Genomic DNA of ZAS-9 yielded only nine fragments upon digestion with PmeI, including a doublet of 63 and 72 kb that under some PFGE conditions appeared as a single band with the greatest mobility (Fig. 2). By contrast, genomic DNA of ZAS-1 yielded 29 fragments with PmeI, even though its genome was 440 kb smaller than that of ZAS-9. Genomes of strain ZAS-2 and S. aurantia J1 also yielded numerous fragments, and many of these occurred as clusters of similar-size fragments that were resolved only after multiple runs under different PFGE conditions. Such clusters appeared as conspicuously bright single bands in some gels (e.g., Fig. 2, lanes 1 to 6).
With the exception of G+C content, S. aurantia genomic DNA was similar in size to that of the termite gut treponemes, and it also contained 2 copies of rrs. It is noteworthy that the G+C content of S. aurantia DNA, determined here directly by HPLC analysis of enzymatically liberated deoxynucleosides (65.6 mol%), was similar to that inferred previously by using the indirect method of equilibrium density centrifugation (ρCsCl): 64.3 mol% (6). However, both of these estimates are substantially higher than that inferred by using the thermal denaturation (Tm) method: 60.0 mol% (6). A discrepancy in G+C content of 4.1 to 5.8 mol%, with the Tm method yielding values consistently lower than those obtained by the ρCsCl method, has been observed for DNAs from other free-living spirochetes as well, e.g., S. stenostrepta and Spirochaeta litoralis (6), but the basis for this discrepancy has not yet been resolved. In this regard, modified deoxynucleosides were not observed during HPLC analysis of DNAs from S. aurantia or any ZAS strain.
The possession of a relatively low rrs copy number by the spirochetes examined here is not surprising, given the average copy number of 1.8 for a range of spirochetes reported in the Ribosomal Operon Copy Number Database (16). The low copy number suggests that the fitness of spirochetes may be tied to an ecological strategy characterized by efficient allocation of resources under relatively constant, slow-growth conditions (15). For termite gut treponemes in particular, this would seem to be entirely consistent with their relatively long doubling times in vitro (Table 1) and with their adaptation to existence in the gut of an animal that feeds on a relatively refractory, N-poor food resource.
Taxonomy of termite gut treponemes.
Treponema strain ZAS-9 is readily distinguished from strains ZAS-1 and ZAS-2 by its cell morphology (Fig. 1), its nonhomoacetogenic fermentation pattern, and the 7 to 8% dissimilarity between its 16S rRNA sequence and those of the latter two treponemes (22). Moreover, under N-limited growth conditions, ZAS-9 exhibits dinitrogen fixation rates at least 10-fold greater than those of ZAS-1 and ZAS-2 and displays unambiguous N2-dependent growth, suggesting that ZAS-9 plays a more important role in supplying fixed N to termites than do ZAS-1 and ZAS-2 (22). We believe that these differences are sufficient to consider ZAS-9 a species distinct from that accommodating ZAS-1 and ZAS-2, and we propose that ZAS-9 be considered the type strain of the new species T. azotonutricium.
By contrast, strains ZAS-1 and ZAS-2 are virtually indistinguishable morphologically; they are both H2-utilizing, CO2-reducing homoacetogens; they are generally similar to each other in nutrition and physiology; and they exhibit 98% similarity in their 16S rRNA sequences (11, 19). The genome of ZAS-2 is about 11% larger than that of ZAS-1, and future work may reveal significant differences between these two spirochetes. However, at this time we believe there is no compelling reason to regard them otherwise than as two strains of the same species, for which we propose the new name T. primitia. Formal descriptions of these new species follow.
Description of Treponema azotonutricium sp. nov.
Treponema azotonutricium sp. nov. (a.zo.to.nu.tri′ci.um. N.L. n. azotum (from French azote), nitrogen; L. neut. adj. nutricium, nourishing; N.L. neut. adj. azotonutricium, nourishing with nitrogen [symbiotic dinitrogen fixation]).
Cells 0.2 to 0.3 μm in diameter by 10 to 12 μm long, with a wavelength or body pitch of 1.2 μm. Motile by two periplasmic flagella, inserted at opposite ends of the protoplasmic cylinder. Anaerobe. Catalase negative. Yeast autolysate required for growth. Optimum temperature for growth is 30°C. Energy sources utilized for fermentative growth include glucose, fructose, ribose, xylose, maltose, and cellobiose. Maltose is fermented to acetate, ethanol, CO2, and H2 as major products. Mannitol, arabinose, sucrose, trehalose, glycine, lactate, pyruvate, uric acid, and H2 (plus CO2) are not utilized. Exhibits nitrogenase activity and N2-dependent growth in media low in combined N. Genome is 3,901 kb and contains 50.0 mol% G+C and 2 rrs gene copies. Nucleotide sequence of the 16S rRNA (GenBank accession no. AF320287) places this spirochete within the “termite cluster” of the genus Treponema (22).
Source: hindgut contents of the Pacific dampwood termite Zootermopsis angusticollis (Hagen) (Isoptera: Termopsidae).
Type strain: strain ZAS-9; deposited with the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany (DSM 13862), and the American Type Culture Collection, Manassas, Va. (ATCC BAA-888).
Description of Treponema primitia sp. nov.
Treponema primitia sp. nov. (pri.mi′ti.a. N.L. fem. sing. n. primitia [nominative in apposition], the first fruit [of isolation after long work]).
Cells 0.2 μm in diameter by 3 to 7 μm long, with a wavelength or body pitch of 2.3 μm. Motile by two periplasmic flagella, inserted at opposite ends of the protoplasmic cylinder. Anaerobe. Possesses NADH and NADPH peroxidases but neither catalase nor superoxide dismutase. Optimum temperature for growth is 30°C. Optimum pH for growth is 7.2 (range, 6.5 to 7.8). Homoacetogen. Energy sources used for growth include glucose, maltose, mannitol, xylose, and H2 (plus CO2), which are fermented to acetate as the sole product. Strain ZAS-1 also uses arabinose and cellobiose, whereas strain ZAS-2 can grow slowly by acetogenic demethylation of methoxylated benzenoids (syringate, ferulate, vanillate, and trimethoxybenzoate). Ribose, methanol, formate, CO, lactate, pyruvate, glycine, betaine, and choline are not utilized. Growth by mixotrophy (i.e., simultaneous use of H2 and organic substrates) has been demonstrated. Laboratory-prepared yeast autolysate or certain commercial yeast extracts are required for growth. Folinate (formyltetrahydrofolate) is required for growth of strain ZAS-1, whereas folic acid or folinate is required by strain ZAS-2. Cells possess homologues of the dinitrogenase reductase gene nifH and exhibit low levels of nitrogenase activity, but unambiguous N2-dependent growth has not been demonstrated. Genome sizes are 3,461 kb (ZAS-1) and 3,835 kb (ZAS-2); G+C contents of DNA are 51.0 mol% (ZAS-1) and 50.9 mol% (ZAS-2) (by HPLC); each strain possesses 2 rrs gene copies. The 16S rRNA nucleotide sequences of strains ZAS-1 (GenBank accession no. AF093251) and ZAS-2 (GenBank accession no. AF093252) place them within the “termite cluster” of the genus Treponema (22).
Source: hindgut contents of the Pacific dampwood termite Zootermopsis angusticollis (Hagen) (Isoptera: Termopsidae).
Type strain: strain ZAS-2; deposited with the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany (DSM 12427), and the American Type Culture Collection, Manassas, Va. (ATCC BAA-887).
Acknowledgments
We thank Kwi Kim and Brendan Keough for help with determination of genome sizes and rrs copy numbers and Kristina Stredwick for providing cells of M. xanthus DK1622. We are also grateful to W. B. Whitman for advice on HPLC analysis of the G+C content of DNA and to Hans G. Trüper for help with nomenclature and construction of Latinized specific epithets.
This research was supported by NSF research grants IBN-9709000 and IBN-0114505 to J.A.B.
REFERENCES
- 1.Beckwith, T. D., and S. F. Light. 1927. The spirals within the termite gut for class use. Science 66:656-657. [DOI] [PubMed] [Google Scholar]
- 2.Bergthorsson, U., and H. Ochman. 1995. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J. Bacteriol. 177:5784-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birren, B., and E. Lai. 1993. Pulsed field gel electrophoresis: a practical guide. Academic Press, Inc., San Diego, Calif..
- 4.Breznak, J. A. 2000. Ecology of prokaryotic microbes in the guts of wood- and litter-feeding termites, p. 209-231. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Breznak, J. A. 2002. Phylogenetic diversity and physiology of termite hindgut spirochetes. Integ. Comp. Biol. 42:313-318. [DOI] [PubMed] [Google Scholar]
- 6.Breznak, J. A., and E. Canale-Parola. 1975. Morphology and physiology of Spirochaeta aurantia strains isolated from aquatic habitats. Arch. Microbiol. 105:1-12. [DOI] [PubMed] [Google Scholar]
- 7.Breznak, J. A., and E. Canale-Parola. 1969. Spirochaeta aurantia, a pigmented, facultatively anaerobic spirochete. J. Bacteriol. 97:386-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breznak, J. A., and J. R. Leadbetter. 2002. Termite gut spirochetes. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. [Online.] Springer-Verlag, New York, N.Y.
- 9.Canale-Parola, E. 1984. Family I. Spirochaetaceae Swellengrebel 1907, 581AL, p. 39-46. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 10.Fraser, C. M., S. J. Norris, C. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, C. Fujii, S. Garland, B. Hatch, K. Horst, K. Roberts, M. Sandusky, J. Weidman, H. O. Smith, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 11.Graber, J. R., and J. A. Breznak. 2004. Physiology and nutrition of Treponema primitia, an H2/CO2-acetogenic spirochete from termite hindguts. Appl. Environ. Microbiol. 70:1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harwood, C. S., and E. Canale-Parola. 1981. Branched-chain amino acid fermentation by a marine spirochete: strategy for starvation survival. J. Bacteriol. 148:109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hespell, R. B., and E. Canale-Parola. 1970. Carbohydrate metabolism in Spirochaeta stenostrepta. J. Bacteriol. 103:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen, P. H., and H. W. Morgan. 1992. Glucose catabolism by Spirochaeta thermophila RI 19.B1. J. Bacteriol. 174:2449-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klappenbach, J. A., and T. M. Schmidt. 11 October 2002, posting date. rrndb, release 2.4. [Online.] Thomas M. Schmidt laboratory, Department of Microbiology and Molecular Genetics, Michigan State University, East Lansing, Mich. http://rrndb.cme.msu.edu/rrndb/servlet/controller.
- 18.Leadbetter, J. R., and J. A. Breznak. 1996. Physiological ecology of Methanobrevibacter cuticularis sp. nov. and Methanobrevibacter curvatus sp. nov., isolated from the hindgut of the termite Reticulitermes flavipes. Appl. Environ. Microbiol. 62:3620-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 20.Leidy, J. 1877. On the intestinal parasites of Termes flavipes. Proc. Acad. Natl. Sci. (Philadelphia) 29:146-149. [Google Scholar]
- 21.Leidy, J. 1874-1881. The parasites of the termites. J. Acad. Natl. Sci. (Philadelphia) 8:425-447. [Google Scholar]
- 22.Lilburn, T. G., K. S. Kim, N. E. Ostrom, K. R. Byzek, J. R. Leadbetter, and J. A. Breznak. 2001. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495-2498. [DOI] [PubMed] [Google Scholar]
- 23.Lilburn, T. G., T. M. Schmidt, and J. A. Breznak. 1999. Phylogenetic diversity of termite gut spirochaetes. Environ. Microbiol. 1:331-345. [DOI] [PubMed] [Google Scholar]
- 24.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 25.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell. Sinauer Associates, Sunderland, Mass.
- 26.Pfennig, N., and S. Wagener. 1986. An improved method of preparing wet mounts for photomicrographs of microorganisms. J. Microbiol. Methods 4:303-306. [Google Scholar]
- 27.Segel, I. H. 1976. Biochemical calculations, 2nd ed. John Wiley & Sons, New York, N.Y.
- 28.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 29.Stanton, T. B. 1989. Glucose metabolism and NADH recycling by Treponema hyodysenteriae, the agent of swine dysentery. Appl. Environ. Microbiol. 55:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stouthamer, A. H. 1979. The search for correlation between theoretical and experimental growth yields. Int. Rev. Biochem. 21:1-47. [Google Scholar]
- 31.Thauer, R. K., K. Jungermann, and K. Decker. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinstock, G. 31 December 2002, revision date. T. denticola Genome Project. [Online.] Human Genome Sequencing Center, Baylor College of Medicine, Houston, Tex. http://hgsc.bcm.tmc.edu/microbial/Tdenticola.
- 33.Zhilina, T. N., G. A. Zavarzin, F. A. Rainey, V. V. Kevbrin, N. A. Kostrikina, and A. M. Lysenko. 1996. Spirochaeta alkalica sp. nov., Spirochaeta africana sp. nov., and Spirochaeta asiatica sp. nov., alkaliphilic anaerobes from the continental soda lakes in Central Asia and the East African Rift. Int. J. Syst. Bacteriol. 46:305-312. [DOI] [PubMed] [Google Scholar]