Fig. 4.
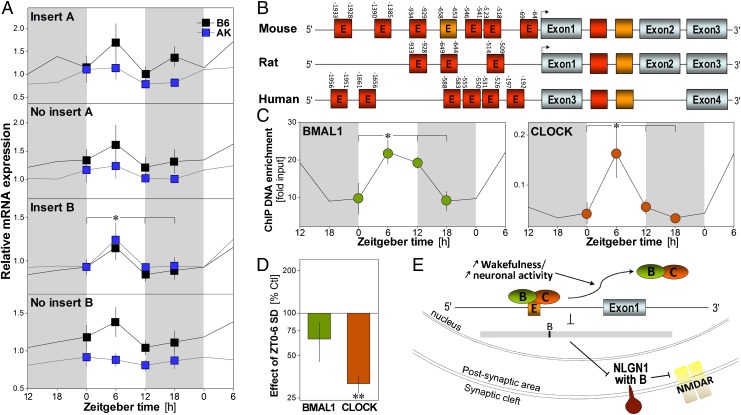
Rhythmic expression of Nlg1 and binding of its gene by clock transcription factors. (A) Relative expression of Nlg1 transcripts measured at ZT0, -6, -12, and -18 in the forebrain of C57BL/6J (B6) and AKR/J (AK) mice (n = 5–8/group). The expression of Nlg1 containing insert B is highest at ZT6 (time effect: F3,49 = 11.7, P < 0.01). A significant strain effect was observed for Nlg1 with A and without B (F3,49 ≥ 4.4, P < 0.05). Gray backgrounds indicate dark periods. *P < 0.05 between indicated values (also in C). (B) Diagram of the mouse, rat, and human Nlg1 gene showing the position of E-boxes relative to the transcription start sites (arrows). Red E-boxes represent CANNTG and orange CACGTG. Absence of E in the box indicates the presence of multiple E-boxes. (C) Binding of BMAL1 and CLOCK to the CACGTG of Nlg1 in B6 mouse cerebral cortex measured at ZT0, -6, -12, and -18 (n = 3/point, except n = 2 for ZT12). BMAL1 and CLOCK bind to Nlg1 in a time-of-day–dependent manner (F3,7 ≥ 4.2, P = 0.05). (D) Binding measured at ZT6 after 6 h of SD and expressed relative to the binding in undisturbed control (n = 5/group). SD decreased CLOCK binding to Nlg1 (t = −15.1, **P < 0.001), whereas a tendency was observed for BMAL1 (t = −1.7, P = 0.17). (E) Model showing how elevated sleep pressure may change Nlg1 expression: prolonged wakefulness could lead to detachment of CLOCK/BMAL1 from the gene, which would decrease expression of Nlg1 with insert B and synaptic NLG1 and down-regulate NMDAR function.